Descriptor analysis/Coarse-graining
Warning
The following procedures and analyses should be considered experimental. They are not well-tested and there is no related peer-reviewed publication.
This short tutorial shows how to prepare and run a descriptor analysis which can
be useful when creating a coarse-grained (CG) model based on neural network
potentials. The steps outlined here correspond to the subfolders of the
examples/coarse-graining directory in the repository. The descriptor
analysis itself is described in detail inside the Jupyter notebook
step-3_cluster-analysis/analyze-descriptors.ipynb.
Note
The tools nnp-atomenv and nnp-scaling need to be compiled to follow
the steps presented in this tutorial.
Step 0 (optional): Coarse-grained data set
Note
This step is optional, the descriptor analysis may also yield insights if applied directly to the full, atomistic description of the system.
First, we need to create a data set which contains the coarse-grained
description of the original structures. In the example (directory
step-0_cg-data-set) we start with a data set of bulk water (input.data)
where we apply the following coarse-graining strategy: for each oxygen atom we
identify the two closest hydrogen neighbors and compute then the center of mass
coordinates of all molecules via
where \(\Delta \vec{r}_{jk} = \vec{r}_j - \vec{r}_k\). The force acting on the center of mass is then just the sum of all forces in each molecule.
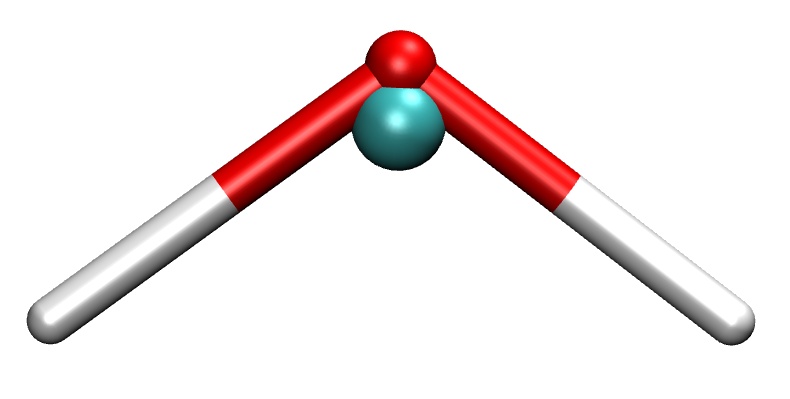
A water molecule and its coarse-grained (center of mass) representation (turquoise sphere).
An example C++ code is provided to perform this conversion in nnp-cgdata.cpp
making use of the libnnp library. To build the executable just run make
(libnnp needs to be compiled beforehand in the main src directory). Then
run the executable with an appropriate cutoff radius for the neighbor list as
the only argument:
./nnp-cgdata 2.3
Here, we use a cutoff of 2.3 bohr which ensures that the two closest hydrogen
neighbors are found for each oxygen atom. This will create the file
output.data which contains the desired coarse-grained center of mass
particles representing the water molecules.
Hint
The “element symbol” of the CG particles in this example is Cm. Normally,
this would correspond to the element Curium but here we use it to abbreviate
“center of mass”.
The nnp-cgdata.cpp code demonstrates the usage of the C++ API to manipulate
datasets of configurations. Of course such a task can also be performed with
external software.
Step 1: Create symmetry function set
This step is common to regular NNP training with n2p2: for a given data set we
need to prepare an appropriate symmetry function set and compute the scaling
data. Please see also this section for further
information. For this example, an input.nn file is already prepared in
step-1_scaling-data which contains symmetry function definitions for our
coarse-grained description of water:
...
symfunction_short Cm 2 Cm 0.30 4.0 12.00
symfunction_short Cm 2 Cm 0.60 4.0 12.00
symfunction_short Cm 2 Cm 1.50 4.0 12.00
symfunction_short Cm 3 Cm Cm 0.03 1.0 1.0 12.00000
symfunction_short Cm 3 Cm Cm 0.03 -1.0 1.0 12.00000
...
Together with the data set file input.data (file is provided, it is the
output.data of step 0 above) we can proceed to compute the symmetry
functions statistics:
mpirun -np 4 ../../../bin/nnp-scaling 500
We can safely ignore (or delete) the output files sf.???.????.histo, we only
require the scaling.data file for the next steps.
Step 2: Generate atomic environment data
Next, in order to feed the descriptor analysis Jupyter notebook in the final
step, we need to prepare atomic environment data for our system. Please read the
description of the tool nnp-atomenv for a detailed
explanation how this data is constructed. The required files from the previous
steps are prepared in the step-2_atomic-env directory. For the
coarse-graining example we use the command
../../../bin/nnp-atomenv 500 4
where the second argument determines the maximum number of neighbor particles
considered for the atomic environment data. Again, we can ignore all output
files except atomic-env.dGdx which we use in the final step.
Step 3: Analyze descriptors
Finally, everything is ready to start the Jupyter notebook
analyze-descriptors.ipynb which carries out the actual descriptor analysis. The
notebook and necessary files are provided in step-3_cluster-analysis. After
starting Jupyter with
jupyter notebook analyze-descriptors.ipynb
we can run all cells which may take a while. All further steps of the descriptor analysis are described in the notebook’s comment cells.