#include <InterfaceLammps.h>
Public Member Functions | |
| InterfaceLammps () | |
| void | initialize (char const *const &directory, char const *const &emap, bool showew, bool resetew, int showewsum, int maxew, double cflength, double cfenergy, double lammpsCutoff, int lammpsNtypes, int myRank) |
| Initialize the LAMMPS interface. | |
| void | setGlobalStructureStatus (bool const status) |
| Specify whether n2p2 knows about global structure or only local structure. | |
| bool | getGlobalStructureStatus () |
| Check if n2p2 knows about global structure. | |
| void | setLocalAtoms (int numAtomsLocal, int const *const atomType) |
| (Re)set structure to contain only local LAMMPS atoms. | |
| void | setLocalAtomPositions (double const *const *const atomPos) |
| Set absolute atom positions from LAMMPS (nnp/develop only). | |
| void | setLocalTags (int const *const atomTag) |
| Set atom tags (int version, -DLAMMPS_SMALLBIG). | |
| void | setLocalTags (int64_t const *const atomTag) |
| Set atom tags (int64_t version, -DLAMMPS_BIGBIG). | |
| void | setBoxVectors (double const *boxlo, double const *boxhi, double const xy, double const xz, double const yz) |
| Set box vectors of structure stored in LAMMPS (nnp/develop only). | |
| void | allocateNeighborlists (int const *const numneigh) |
| Allocate neighbor lists. | |
| void | addNeighbor (int i, int j, int64_t tag, int type, double dx, double dy, double dz, double d2) |
| Add one neighbor to atom (int64_t version, -DLAMMPS_BIGBIG). | |
| void | finalizeNeighborList () |
| Sorts neighbor list and creates cutoff map if necessary. | |
| void | process () |
| Calculate symmetry functions, atomic neural networks and sum of local energy contributions. | |
| void | processDevelop () |
| Calculate symmetry functions, atomic neural networks and sum of local energy contributions (development version for "hdnnp/develop" pair style). | |
| double | getEnergy () const |
| Return sum of local energy contributions. | |
| double | getAtomicEnergy (int index) const |
| Return energy contribution of one atom. | |
| void | addElectrostaticEnergy (double energy) |
| Adds electrostatic energy contribution to the total structure energy. | |
| void | getForces (double *const *const &atomF) const |
| Calculate forces and add to LAMMPS atomic force arrays. | |
| void | getForcesDevelop (double *const *const &atomF) const |
| Calculate forces and add to LAMMPS atomic force arrays (development version for "hdnnp/develop" pair style). | |
| void | getForcesChi (double const *const &lambda, double *const *const &atomF) const |
| Calculate chi-term for forces and add to LAMMPS atomic force arrays. | |
| void | getCharges (double *const &atomQ) const |
| Transfer charges (in units of e) to LAMMPS atomic charge vector. | |
| bool | isInitialized () const |
| Check if this interface is correctly initialized. | |
| double | getMaxCutoffRadius () const |
| Get largest cutoff of symmetry functions. | |
| double | getEwaldPrec () const |
| Get Ewald precision parameter. | |
| double | getMaxCutoffRadiusOverall () |
| Get largest cutoff including structure specific cutoff and screening cutoff. | |
| long | getEWBufferSize () const |
| Calculate buffer size for extrapolation warning communication. | |
| void | fillEWBuffer (char *const &buf, int bs) const |
| Fill provided buffer with extrapolation warning entries. | |
| void | extractEWBuffer (char const *const &buf, int bs) |
| Extract given buffer to symmetry function statistics class. | |
| void | writeExtrapolationWarnings () |
| Write extrapolation warnings to log. | |
| void | clearExtrapolationWarnings () |
| Clear extrapolation warnings storage. | |
| void | addCharge (int index, double Q) |
| Read atomic charges from LAMMPS into n2p2. | |
| void | getQEqParams (double *const &atomChi, double *const &atomJ, double *const &sigmaSqrtPi, double *const *const &gammaSqrt2, double &qRef) const |
| Write QEq arrays from n2p2 to LAMMPS. | |
| void | getdEdQ (double *const &dEtotdQ) const |
| Write the derivative of total energy with respect to atomic charges from n2p2 into LAMMPS. | |
| void | getScreeningInfo (double *const &rScreen) const |
| Read screening function information from n2p2 into LAMMPS. | |
| void | getdChidxyz (int tag, double *const &dChidx, double *const &dChidy, double *const &dChidz) const |
| Transfer spatial derivatives of atomic electronegativities. | |
| void | setElecDone () |
| Set isElecDone true after running the first NN in 4G-HDNNPs. | |
| void | writeToFile (std::string const fileName, bool const append) |
| Write current structure to file in units used in training data. | |
| void | add3DVecToArray (double *const &arr, Vec3D const &v) const |
| Add a Vec3D vector to a 3D array in place. | |
 Public Member Functions inherited from nnp::Mode Public Member Functions inherited from nnp::Mode | |
| Mode () | |
| void | initialize () |
| Write welcome message with version information. | |
| void | loadSettingsFile (std::string const &fileName="input.nn") |
| Open settings file and load all keywords into memory. | |
| void | setupGeneric (std::string const &nnpDir="", bool skipNormalize=false, bool initialHardness=false) |
| Combine multiple setup routines and provide a basic NNP setup. | |
| void | setupNormalization (bool standalone=true) |
| Set up normalization. | |
| virtual void | setupElementMap () |
| Set up the element map. | |
| virtual void | setupElements () |
| Set up all Element instances. | |
| void | setupCutoff () |
| Set up cutoff function for all symmetry functions. | |
| virtual void | setupSymmetryFunctions () |
| Set up all symmetry functions. | |
| void | setupSymmetryFunctionScalingNone () |
| Set up "empy" symmetry function scaling. | |
| virtual void | setupSymmetryFunctionScaling (std::string const &fileName="scaling.data") |
| Set up symmetry function scaling from file. | |
| virtual void | setupSymmetryFunctionGroups () |
| Set up symmetry function groups. | |
| virtual void | setupSymmetryFunctionCache (bool verbose=false) |
| Set up symmetry function cache. | |
| void | setupSymmetryFunctionMemory (bool verbose=false) |
| Extract required memory dimensions for symmetry function derivatives. | |
| void | setupSymmetryFunctionStatistics (bool collectStatistics, bool collectExtrapolationWarnings, bool writeExtrapolationWarnings, bool stopOnExtrapolationWarnings) |
| Set up symmetry function statistics collection. | |
| void | setupCutoffMatrix () |
| Setup matrix storing all symmetry function cut-offs for each element. | |
| virtual void | setupNeuralNetwork () |
| Set up neural networks for all elements. | |
| virtual void | setupNeuralNetworkWeights (std::map< std::string, std::string > fileNameFormats=std::map< std::string, std::string >()) |
| Set up neural network weights from files with given name format. | |
| virtual void | setupNeuralNetworkWeights (std::string directoryPrefix, std::map< std::string, std::string > fileNameFormats=std::map< std::string, std::string >()) |
| Set up neural network weights from files with given name format. | |
| virtual void | setupElectrostatics (bool initialHardness=false, std::string directoryPrefix="", std::string fileNameFormat="hardness.%03zu.data") |
| Set up electrostatics related stuff (hardness, screening, ...). | |
| void | calculateSymmetryFunctions (Structure &structure, bool const derivatives) |
| Calculate all symmetry functions for all atoms in given structure. | |
| void | calculateSymmetryFunctionGroups (Structure &structure, bool const derivatives) |
| Calculate all symmetry function groups for all atoms in given structure. | |
| void | calculateAtomicNeuralNetworks (Structure &structure, bool const derivatives, std::string id="") |
| Calculate a single atomic neural network for a given atom and nn type. | |
| void | chargeEquilibration (Structure &structure, bool const derivativesElec) |
| Perform global charge equilibration method. | |
| void | calculateEnergy (Structure &structure) const |
| Calculate potential energy for a given structure. | |
| void | calculateCharge (Structure &structure) const |
| Calculate total charge for a given structure. | |
| void | calculateForces (Structure &structure) const |
| Calculate forces for all atoms in given structure. | |
| void | evaluateNNP (Structure &structure, bool useForces=true, bool useDEdG=true) |
| Evaluate neural network potential (includes total energy, optionally forces and in some cases charges. | |
| void | addEnergyOffset (Structure &structure, bool ref=true) |
| Add atomic energy offsets to reference energy. | |
| void | removeEnergyOffset (Structure &structure, bool ref=true) |
| Remove atomic energy offsets from reference energy. | |
| double | getEnergyOffset (Structure const &structure) const |
| Get atomic energy offset for given structure. | |
| double | getEnergyWithOffset (Structure const &structure, bool ref=true) const |
| Add atomic energy offsets and return energy. | |
| double | normalized (std::string const &property, double value) const |
| Apply normalization to given property. | |
| double | normalizedEnergy (Structure const &structure, bool ref=true) const |
| Apply normalization to given energy of structure. | |
| double | physical (std::string const &property, double value) const |
| Undo normalization for a given property. | |
| double | physicalEnergy (Structure const &structure, bool ref=true) const |
| Undo normalization for a given energy of structure. | |
| void | convertToNormalizedUnits (Structure &structure) const |
| Convert one structure to normalized units. | |
| void | convertToPhysicalUnits (Structure &structure) const |
| Convert one structure to physical units. | |
| void | logEwaldCutoffs () |
| Logs Ewald params whenever they change. | |
| std::size_t | getNumExtrapolationWarnings () const |
| Count total number of extrapolation warnings encountered for all elements and symmetry functions. | |
| void | resetExtrapolationWarnings () |
| Erase all extrapolation warnings and reset counters. | |
| NNPType | getNnpType () const |
| Getter for Mode::nnpType. | |
| double | getMeanEnergy () const |
| Getter for Mode::meanEnergy. | |
| double | getConvEnergy () const |
| Getter for Mode::convEnergy. | |
| double | getConvLength () const |
| Getter for Mode::convLength. | |
| double | getConvCharge () const |
| Getter for Mode::convCharge. | |
| double | getEwaldPrecision () const |
| Getter for Mode::ewaldSetup.precision. | |
| double | getEwaldMaxCharge () const |
| Getter for Mode::ewaldSetup.maxCharge. | |
| double | getEwaldMaxSigma () const |
| Getter for Mode::ewaldSetup.maxQsigma. | |
| EWALDTruncMethod | getEwaldTruncationMethod () const |
| Getter for Mode::ewaldSetup.truncMethod. | |
| KSPACESolver | kspaceSolver () const |
| Getter for Mode::kspaceSolver. | |
| double | getMaxCutoffRadius () const |
| Getter for Mode::maxCutoffRadius. | |
| std::size_t | getNumElements () const |
| Getter for Mode::numElements. | |
| ScreeningFunction | getScreeningFunction () const |
| Getter for Mode::screeningFunction. | |
| std::vector< std::size_t > | getNumSymmetryFunctions () const |
| Get number of symmetry functions per element. | |
| bool | useNormalization () const |
| Check if normalization is enabled. | |
| bool | settingsKeywordExists (std::string const &keyword) const |
| Check if keyword was found in settings file. | |
| std::string | settingsGetValue (std::string const &keyword) const |
| Get value for given keyword in Settings instance. | |
| std::vector< std::size_t > | pruneSymmetryFunctionsRange (double threshold) |
| Prune symmetry functions according to their range and write settings file. | |
| std::vector< std::size_t > | pruneSymmetryFunctionsSensitivity (double threshold, std::vector< std::vector< double > > sensitivity) |
| Prune symmetry functions with sensitivity analysis data. | |
| void | writePrunedSettingsFile (std::vector< std::size_t > prune, std::string fileName="output.nn") const |
| Copy settings file but comment out lines provided. | |
| void | writeSettingsFile (std::ofstream *const &file) const |
| Write complete settings file. | |
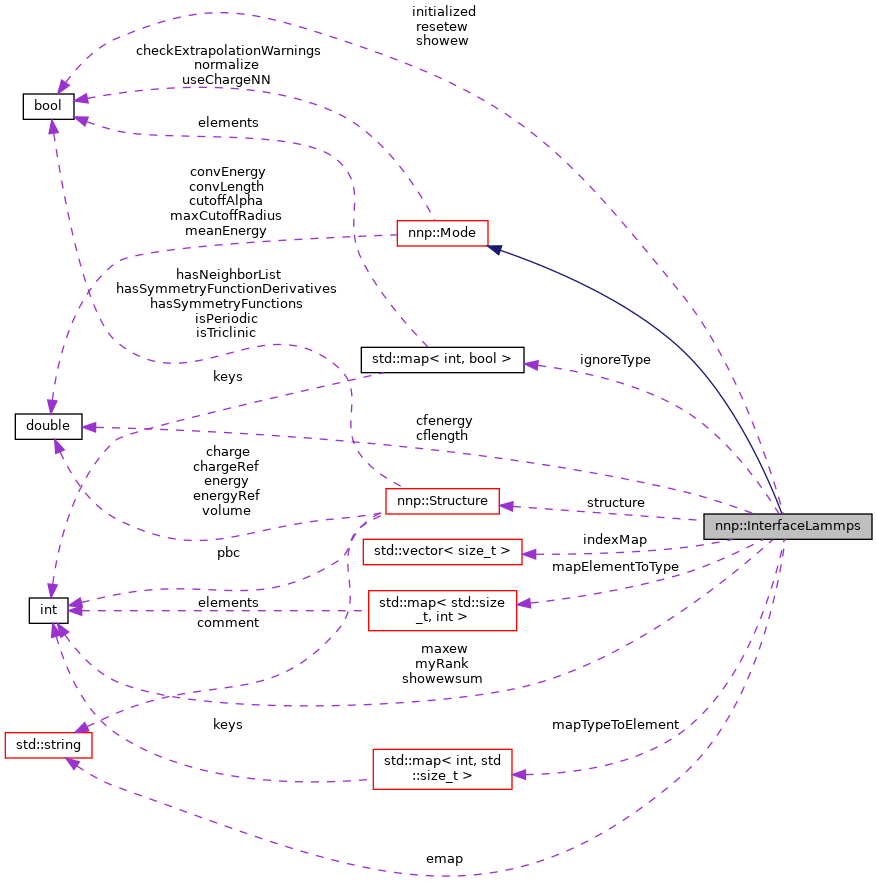
Protected Attributes | |
| int | myRank |
| Process rank. | |
| bool | initialized |
| Initialization state. | |
| bool | hasGlobalStructure |
| Whether n2p2 knows about the global structure or only a local part. | |
| bool | showew |
Corresponds to LAMMPS showew keyword. | |
| bool | resetew |
Corresponds to LAMMPS resetew keyword. | |
| int | showewsum |
Corresponds to LAMMPS showewsum keyword. | |
| int | maxew |
Corresponds to LAMMPS maxew keyword. | |
| double | cflength |
Corresponds to LAMMPS cflength keyword. | |
| double | cfenergy |
Corresponds to LAMMPS cfenergy keyword. | |
| std::string | emap |
Corresponds to LAMMPS map keyword. | |
| std::vector< size_t > | indexMap |
| Map from LAMMPS index to n2p2 atom index. | |
| std::map< int, bool > | ignoreType |
| True if atoms of this LAMMPS type will be ignored. | |
| std::map< int, std::size_t > | mapTypeToElement |
| Map from LAMMPS type to n2p2 element index. | |
| std::map< std::size_t, int > | mapElementToType |
| Map from n2p2 element index to LAMMPS type. | |
| Structure | structure |
| Structure containing local atoms. | |
| bool | isElecDone |
| True if first NN is calculated. | |
 Protected Attributes inherited from nnp::Mode Protected Attributes inherited from nnp::Mode | |
| NNPType | nnpType |
| bool | normalize |
| bool | checkExtrapolationWarnings |
| std::size_t | numElements |
| std::vector< std::size_t > | minNeighbors |
| std::vector< double > | minCutoffRadius |
| double | maxCutoffRadius |
| double | cutoffAlpha |
| double | meanEnergy |
| double | convEnergy |
| double | convLength |
| double | convCharge |
| double | fourPiEps |
| EwaldSetup | ewaldSetup |
| KspaceGrid | kspaceGrid |
| settings::Settings | settings |
| SymFnc::ScalingType | scalingType |
| CutoffFunction::CutoffType | cutoffType |
| ScreeningFunction | screeningFunction |
| std::vector< Element > | elements |
| std::vector< std::string > | nnk |
| std::map< std::string, NNSetup > | nns |
| std::vector< std::vector< double > > | cutoffs |
| Matrix storing all symmetry function cut-offs for all elements. | |
| ErfcBuf | erfcBuf |
Additional Inherited Members | |
 Public Types inherited from nnp::Mode Public Types inherited from nnp::Mode | |
| enum class | NNPType { HDNNP_2G = 2 , HDNNP_4G = 4 , HDNNP_Q = 10 } |
 Public Attributes inherited from nnp::Mode Public Attributes inherited from nnp::Mode | |
| ElementMap | elementMap |
| Global element map, populated by setupElementMap(). | |
| Log | log |
| Global log file. | |
 Protected Member Functions inherited from nnp::Mode Protected Member Functions inherited from nnp::Mode | |
| void | readNeuralNetworkWeights (std::string const &id, std::string const &fileName) |
| Read in weights for a specific type of neural network. | |
Detailed Description
Definition at line 31 of file InterfaceLammps.h.
Constructor & Destructor Documentation
◆ InterfaceLammps()
| InterfaceLammps::InterfaceLammps | ( | ) |
Definition at line 38 of file InterfaceLammps.cpp.
References cfenergy, cflength, hasGlobalStructure, initialized, isElecDone, maxew, myRank, resetew, showew, and showewsum.
Member Function Documentation
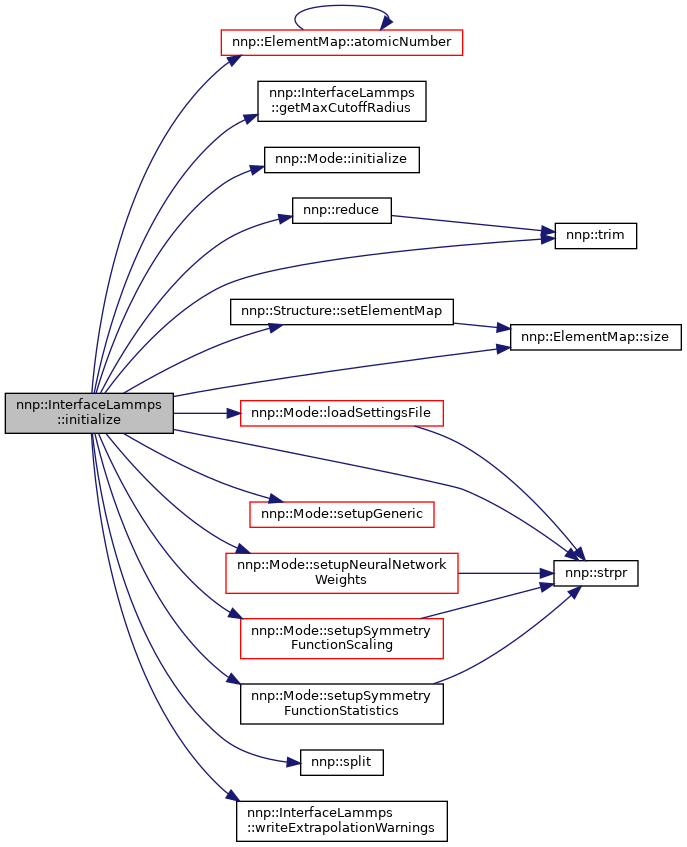
◆ initialize()
| void InterfaceLammps::initialize | ( | char const *const & | directory, |
| char const *const & | emap, | ||
| bool | showew, | ||
| bool | resetew, | ||
| int | showewsum, | ||
| int | maxew, | ||
| double | cflength, | ||
| double | cfenergy, | ||
| double | lammpsCutoff, | ||
| int | lammpsNtypes, | ||
| int | myRank ) |
Initialize the LAMMPS interface.
- Parameters
-
[in] directory Directory containing NNP data files (weights, scaling, settings). [in] emap Element mapping from LAMMPS to n2p2. [in] showew If detailed extrapolation warnings for all atoms are shown. [in] resetew If extrapolation warnings counter is reset every timestep. [in] showewsum Show number of warnings every this many timesteps. [in] maxew Abort simulation if more than this many warnings are encountered. [in] cflength Length unit conversion factor. [in] cfenergy Energy unit conversion factor. [in] lammpsCutoff Cutoff radius from LAMMPS (via pair_coeff). [in] lammpsNtypes Number of atom types in LAMMPS. [in] myRank MPI process rank (passed on to structure index).
Definition at line 51 of file InterfaceLammps.cpp.
References cfenergy, cflength, nnp::Mode::elementMap, emap, getMaxCutoffRadius(), ignoreType, nnp::Mode::initialize(), initialized, nnp::Mode::loadSettingsFile(), nnp::Mode::log, mapElementToType, mapTypeToElement, maxew, myRank, nnp::reduce(), resetew, nnp::Mode::setupGeneric(), nnp::Mode::setupNeuralNetworkWeights(), nnp::Mode::setupSymmetryFunctionScaling(), nnp::Mode::setupSymmetryFunctionStatistics(), showew, showewsum, nnp::split(), nnp::strpr(), structure, TOLCUTOFF, nnp::trim(), and writeExtrapolationWarnings().
◆ setGlobalStructureStatus()
| void InterfaceLammps::setGlobalStructureStatus | ( | bool const | status | ) |
Specify whether n2p2 knows about global structure or only local structure.
- Parameters
-
[in] status true if n2p2 has global structure.
Definition at line 257 of file InterfaceLammps.cpp.
References hasGlobalStructure.
◆ getGlobalStructureStatus()
| bool InterfaceLammps::getGlobalStructureStatus | ( | ) |
Check if n2p2 knows about global structure.
Definition at line 262 of file InterfaceLammps.cpp.
References hasGlobalStructure.
◆ setLocalAtoms()
| void InterfaceLammps::setLocalAtoms | ( | int | numAtomsLocal, |
| int const *const | atomType ) |
(Re)set structure to contain only local LAMMPS atoms.
- Parameters
-
[in] numAtomsLocal Number of local atoms. [in] atomType LAMMPS atom type.
Definition at line 267 of file InterfaceLammps.cpp.
References nnp::Atom::element, nnp::Atom::hasSymmetryFunctionDerivatives, nnp::Atom::hasSymmetryFunctions, ignoreType, nnp::Atom::index, indexMap, nnp::Atom::indexStructure, mapTypeToElement, myRank, nnp::Atom::neighbors, nnp::Mode::numElements, nnp::Atom::numNeighbors, nnp::Atom::numNeighborsPerElement, and structure.
◆ setLocalAtomPositions()
| void InterfaceLammps::setLocalAtomPositions | ( | double const *const *const | atomPos | ) |
Set absolute atom positions from LAMMPS (nnp/develop only).
- Parameters
-
[in] atomPos Atom coordinate array in LAMMPS units.
Definition at line 306 of file InterfaceLammps.cpp.
References cflength, nnp::Mode::convLength, nnp::Mode::normalize, nnp::Atom::r, and structure.
◆ setLocalTags() [1/2]
| void InterfaceLammps::setLocalTags | ( | int const *const | atomTag | ) |
Set atom tags (int version, -DLAMMPS_SMALLBIG).
- Parameters
-
[in] atomTag LAMMPS atom tag.
Definition at line 325 of file InterfaceLammps.cpp.
References structure.
◆ setLocalTags() [2/2]
| void InterfaceLammps::setLocalTags | ( | int64_t const *const | atomTag | ) |
Set atom tags (int64_t version, -DLAMMPS_BIGBIG).
- Parameters
-
[in] atomTag LAMMPS atom tag.
Definition at line 336 of file InterfaceLammps.cpp.
References structure.
◆ setBoxVectors()
| void InterfaceLammps::setBoxVectors | ( | double const * | boxlo, |
| double const * | boxhi, | ||
| double const | xy, | ||
| double const | xz, | ||
| double const | yz ) |
Set box vectors of structure stored in LAMMPS (nnp/develop only).
- Parameters
-
[in] boxlo Array containing coordinates of origin xlo, ylo, zlo. [in] boxhi Array containing coordinates xhi, yhi, zhi. [in] xy Tilt factor for box vector b. [in] xz First tilt factor for box vector c. [in] yz Second tilt factor for box vector c.
Definition at line 346 of file InterfaceLammps.cpp.
References cflength, nnp::Mode::convLength, nnp::Mode::normalize, and structure.
◆ allocateNeighborlists()
| void InterfaceLammps::allocateNeighborlists | ( | int const *const | numneigh | ) |
Allocate neighbor lists.
- Parameters
-
[in] numneigh Array containing number of neighbors for each local atom.
Definition at line 401 of file InterfaceLammps.cpp.
References structure.
◆ addNeighbor()
| void InterfaceLammps::addNeighbor | ( | int | i, |
| int | j, | ||
| int64_t | tag, | ||
| int | type, | ||
| double | dx, | ||
| double | dy, | ||
| double | dz, | ||
| double | d2 ) |
Add one neighbor to atom (int64_t version, -DLAMMPS_BIGBIG).
- Parameters
-
[in] i Local atom index. [in] j Neighbor atom index. [in] tag Neighbor atom tag. [in] type Neighbor atom type. [in] dx Neighbor atom distance in x direction. [in] dy Neighbor atom distance in y direction. [in] dz Neighbor atom distance in z direction. [in] d2 Square of neighbor atom distance.
If -DLAMMPS_SMALLBIG implicit conversion is applied for tag.
Definition at line 410 of file InterfaceLammps.cpp.
References cflength, nnp::Mode::convLength, nnp::Atom::Neighbor::d, nnp::Atom::Neighbor::dr, nnp::Atom::Neighbor::element, ignoreType, nnp::Atom::Neighbor::index, indexMap, mapTypeToElement, nnp::Atom::neighbors, nnp::Mode::normalize, nnp::Atom::numNeighbors, nnp::Atom::numNeighborsPerElement, structure, and nnp::Atom::Neighbor::tag.
◆ finalizeNeighborList()
| void InterfaceLammps::finalizeNeighborList | ( | ) |
Sorts neighbor list and creates cutoff map if necessary.
If structure is periodic, this function needs to be called after setBoxVectors!
Definition at line 443 of file InterfaceLammps.cpp.
References nnp::Mode::cutoffs, nnp::Mode::ewaldSetup, nnp::Mode::HDNNP_4G, nnp::Mode::maxCutoffRadius, nnp::Mode::nnpType, nnp::Mode::screeningFunction, and structure.
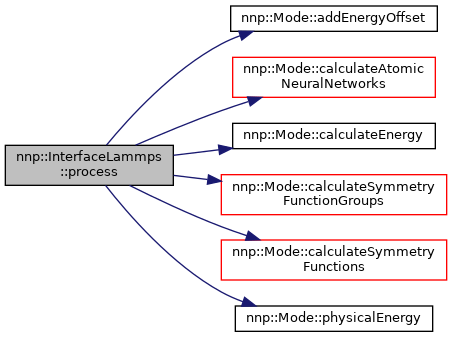
◆ process()
| void InterfaceLammps::process | ( | ) |
Calculate symmetry functions, atomic neural networks and sum of local energy contributions.
Definition at line 463 of file InterfaceLammps.cpp.
References nnp::Mode::addEnergyOffset(), nnp::Mode::calculateAtomicNeuralNetworks(), nnp::Mode::calculateEnergy(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), nnp::Mode::HDNNP_2G, nnp::Mode::HDNNP_4G, isElecDone, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Mode::physicalEnergy(), and structure.
◆ processDevelop()
| void InterfaceLammps::processDevelop | ( | ) |
Calculate symmetry functions, atomic neural networks and sum of local energy contributions (development version for "hdnnp/develop" pair style).
Definition at line 502 of file InterfaceLammps.cpp.
References nnp::Mode::addEnergyOffset(), nnp::Mode::calculateAtomicNeuralNetworks(), nnp::Mode::calculateCharge(), nnp::Mode::calculateEnergy(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), cflength, nnp::Mode::chargeEquilibration(), nnp::Mode::convLength, nnp::Mode::ewaldSetup, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Mode::log, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Mode::physicalEnergy(), and structure.
◆ getEnergy()
| double InterfaceLammps::getEnergy | ( | ) | const |
◆ getAtomicEnergy()
| double InterfaceLammps::getAtomicEnergy | ( | int | index | ) | const |
Return energy contribution of one atom.
- Parameters
-
[in] index Atom index.
- Returns
- energy contribution of atom with given index.
- Attention
- These atomic contributions are not physical!
Definition at line 555 of file InterfaceLammps.cpp.
References cfenergy, nnp::Atom::element, nnp::Mode::elements, nnp::Atom::energy, nnp::Element::getAtomicEnergyOffset(), nnp::Mode::meanEnergy, nnp::Mode::normalize, nnp::Mode::physical(), and structure.
◆ addElectrostaticEnergy()
| void InterfaceLammps::addElectrostaticEnergy | ( | double | energy | ) |
Adds electrostatic energy contribution to the total structure energy.
- Parameters
-
[in] electrostatic energy (calculated in LAMMPS).
Definition at line 674 of file InterfaceLammps.cpp.
References structure.
◆ getForces()
| void InterfaceLammps::getForces | ( | double *const *const & | atomF | ) | const |
Calculate forces and add to LAMMPS atomic force arrays.
- Parameters
-
[in,out] atomF LAMMPS force array for local and ghost atoms.
Definition at line 679 of file InterfaceLammps.cpp.
References cfenergy, cflength, nnp::Mode::convEnergy, nnp::Mode::convLength, nnp::Atom::dEdG, nnp::Atom::dGdr, nnp::Atom::element, nnp::Mode::elements, nnp::Atom::index, nnp::Atom::neighbors, nnp::Mode::normalize, nnp::Atom::numSymmetryFunctions, and structure.
◆ getForcesDevelop()
| void InterfaceLammps::getForcesDevelop | ( | double *const *const & | atomF | ) | const |
Calculate forces and add to LAMMPS atomic force arrays (development version for "hdnnp/develop" pair style).
- Parameters
-
[in,out] atomF LAMMPS force array for local and ghost atoms.
Definition at line 742 of file InterfaceLammps.cpp.
References add3DVecToArray(), nnp::Structure::atoms, nnp::Structure::calculateForceLambdaTotal(), nnp::Atom::calculatePairForceShort(), nnp::Atom::calculateSelfForceShort(), cfenergy, cflength, nnp::Mode::convEnergy, nnp::Mode::convLength, nnp::Atom::element, nnp::Mode::elements, nnp::Atom::getStoredMinNumNeighbors(), nnp::Mode::HDNNP_4G, nnp::Atom::index, nnp::Atom::Neighbor::index, nnp::Mode::maxCutoffRadius, nnp::Atom::neighbors, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Structure::numAtoms, and structure.
◆ getForcesChi()
| void InterfaceLammps::getForcesChi | ( | double const *const & | lambda, |
| double *const *const & | atomF ) const |
Calculate chi-term for forces and add to LAMMPS atomic force arrays.
- Parameters
-
[in,out] atomF LAMMPS force array for local and ghost atoms.
Definition at line 832 of file InterfaceLammps.cpp.
References cfenergy, cflength, nnp::Mode::convEnergy, nnp::Mode::convLength, nnp::Atom::dChidG, nnp::Atom::dGdr, nnp::Atom::element, nnp::Mode::elements, nnp::Atom::index, nnp::Atom::neighbors, nnp::Mode::normalize, nnp::Atom::numSymmetryFunctions, structure, and nnp::Atom::tag.
◆ getCharges()
| void InterfaceLammps::getCharges | ( | double *const & | atomQ | ) | const |
Transfer charges (in units of e) to LAMMPS atomic charge vector.
Call after getAtomicEnergy().
- Parameters
-
[in,out] atomQ LAMMPS charge vector.
Definition at line 903 of file InterfaceLammps.cpp.
References nnp::Structure::atoms, nnp::Mode::HDNNP_4G, nnp::Mode::nnpType, nnp::Structure::numAtoms, and structure.
◆ isInitialized()
|
inline |
Check if this interface is correctly initialized.
- Returns
Trueif initialized,Falseotherwise.
Definition at line 326 of file InterfaceLammps.h.
References initialized.
◆ getMaxCutoffRadius()
| double InterfaceLammps::getMaxCutoffRadius | ( | ) | const |
Get largest cutoff of symmetry functions.
- Returns
- Largest cutoff of all symmetry functions.
Definition at line 528 of file InterfaceLammps.cpp.
References cflength, nnp::Mode::convLength, nnp::Mode::maxCutoffRadius, and nnp::Mode::normalize.
Referenced by getMaxCutoffRadiusOverall(), and initialize().

◆ getEwaldPrec()
| double InterfaceLammps::getEwaldPrec | ( | ) | const |
Get Ewald precision parameter.
- Returns
- Ewald precision parameter.
Definition at line 664 of file InterfaceLammps.cpp.
References nnp::Mode::getEwaldPrecision().
◆ getMaxCutoffRadiusOverall()
| double InterfaceLammps::getMaxCutoffRadiusOverall | ( | ) |
Get largest cutoff including structure specific cutoff and screening cutoff.
- Returns
- Largest cutoff of all symmetry functions and structure specific cutoff and screening cutoff.
Definition at line 534 of file InterfaceLammps.cpp.
References cflength, nnp::Mode::convLength, nnp::Mode::ewaldSetup, getMaxCutoffRadius(), nnp::Mode::HDNNP_4G, nnp::Mode::maxCutoffRadius, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Mode::screeningFunction, and structure.
◆ getEWBufferSize()
| long InterfaceLammps::getEWBufferSize | ( | ) | const |
Calculate buffer size for extrapolation warning communication.
- Returns
- Buffer size.
Definition at line 920 of file InterfaceLammps.cpp.
References nnp::Mode::elements, and MPI_SIZE_T.
◆ fillEWBuffer()
| void InterfaceLammps::fillEWBuffer | ( | char *const & | buf, |
| int | bs ) const |
Fill provided buffer with extrapolation warning entries.
- Parameters
-
[in,out] buf Communication buffer to fill. [in] bs Buffer size.
Definition at line 957 of file InterfaceLammps.cpp.
References nnp::Mode::elements, MPI_SIZE_T, and p.
◆ extractEWBuffer()
| void InterfaceLammps::extractEWBuffer | ( | char const *const & | buf, |
| int | bs ) |
Extract given buffer to symmetry function statistics class.
- Parameters
-
[in] buf Buffer with extrapolation warnings data. [in] bs Buffer size.
Definition at line 990 of file InterfaceLammps.cpp.
References d, nnp::Mode::elements, MPI_SIZE_T, and p.
◆ writeExtrapolationWarnings()
| void InterfaceLammps::writeExtrapolationWarnings | ( | ) |
Write extrapolation warnings to log.
Definition at line 1033 of file InterfaceLammps.cpp.
References nnp::Mode::elements, and nnp::Mode::log.
Referenced by initialize().
◆ clearExtrapolationWarnings()
| void InterfaceLammps::clearExtrapolationWarnings | ( | ) |
Clear extrapolation warnings storage.
Definition at line 1049 of file InterfaceLammps.cpp.
References nnp::Mode::elements.
◆ addCharge()
| void InterfaceLammps::addCharge | ( | int | index, |
| double | Q ) |
Read atomic charges from LAMMPS into n2p2.
Definition at line 648 of file InterfaceLammps.cpp.
References nnp::Atom::charge, and structure.
◆ getQEqParams()
| void InterfaceLammps::getQEqParams | ( | double *const & | atomChi, |
| double *const & | atomJ, | ||
| double *const & | sigmaSqrtPi, | ||
| double *const *const & | gammaSqrt2, | ||
| double & | qRef ) const |
Write QEq arrays from n2p2 to LAMMPS.
- Parameters
-
[in] atomChi Electronegativities. [in] atomJ Atomic hardness. [in] atomSigma Gaussian width. [in] qRef Reference charge of the structure.
Definition at line 572 of file InterfaceLammps.cpp.
References nnp::Atom::chi, nnp::Mode::elements, nnp::Atom::index, nnp::Mode::numElements, and structure.
◆ getdEdQ()
| void InterfaceLammps::getdEdQ | ( | double *const & | dEtotdQ | ) | const |
Write the derivative of total energy with respect to atomic charges from n2p2 into LAMMPS.
- Parameters
-
[in] dEtotdQ Derivative of the total energy w.r.t. atomic charge.
Definition at line 595 of file InterfaceLammps.cpp.
References nnp::Atom::dEdG, nnp::Atom::index, and structure.
◆ getScreeningInfo()
| void InterfaceLammps::getScreeningInfo | ( | double *const & | rScreen | ) | const |
Read screening function information from n2p2 into LAMMPS.
- Parameters
-
[in] rScreen Array that contains screening radii.
Definition at line 656 of file InterfaceLammps.cpp.
References nnp::Mode::screeningFunction.
◆ getdChidxyz()
| void InterfaceLammps::getdChidxyz | ( | int | tag, |
| double *const & | dChidx, | ||
| double *const & | dChidy, | ||
| double *const & | dChidz ) const |
Transfer spatial derivatives of atomic electronegativities.
- Parameters
-
[in] tag Atom of interest dChidx dChidy dChidz
Definition at line 606 of file InterfaceLammps.cpp.
References nnp::Atom::dChidG, nnp::Atom::dGdr, nnp::Atom::element, nnp::Mode::elements, nnp::Atom::index, nnp::Mode::maxCutoffRadius, nnp::Atom::neighbors, nnp::Atom::numSymmetryFunctions, and structure.
◆ setElecDone()
| void InterfaceLammps::setElecDone | ( | ) |
Set isElecDone true after running the first NN in 4G-HDNNPs.
Definition at line 669 of file InterfaceLammps.cpp.
References isElecDone.
◆ writeToFile()
| void InterfaceLammps::writeToFile | ( | std::string const | fileName, |
| bool const | append ) |
Write current structure to file in units used in training data.
- Parameters
-
fileName File name of the output structure file. append true if structure should be appended to existing file.
Definition at line 1060 of file InterfaceLammps.cpp.
References nnp::Mode::convCharge, nnp::Mode::convEnergy, nnp::Mode::convLength, nnp::Mode::meanEnergy, and structure.
◆ add3DVecToArray()
| void InterfaceLammps::add3DVecToArray | ( | double *const & | arr, |
| Vec3D const & | v ) const |
Add a Vec3D vector to a 3D array in place.
- Parameters
-
[in,out] arr Array which is edited in place. [in] v Vector which is added to arr.
Definition at line 1068 of file InterfaceLammps.cpp.
Referenced by getForcesDevelop().
Member Data Documentation
◆ myRank
|
protected |
Process rank.
Definition at line 289 of file InterfaceLammps.h.
Referenced by initialize(), InterfaceLammps(), and setLocalAtoms().
◆ initialized
|
protected |
Initialization state.
Definition at line 291 of file InterfaceLammps.h.
Referenced by initialize(), InterfaceLammps(), and isInitialized().
◆ hasGlobalStructure
|
protected |
Whether n2p2 knows about the global structure or only a local part.
Definition at line 293 of file InterfaceLammps.h.
Referenced by getGlobalStructureStatus(), InterfaceLammps(), and setGlobalStructureStatus().
◆ showew
|
protected |
Corresponds to LAMMPS showew keyword.
Definition at line 295 of file InterfaceLammps.h.
Referenced by initialize(), and InterfaceLammps().
◆ resetew
|
protected |
Corresponds to LAMMPS resetew keyword.
Definition at line 297 of file InterfaceLammps.h.
Referenced by initialize(), and InterfaceLammps().
◆ showewsum
|
protected |
Corresponds to LAMMPS showewsum keyword.
Definition at line 299 of file InterfaceLammps.h.
Referenced by initialize(), and InterfaceLammps().
◆ maxew
|
protected |
Corresponds to LAMMPS maxew keyword.
Definition at line 301 of file InterfaceLammps.h.
Referenced by initialize(), and InterfaceLammps().
◆ cflength
|
protected |
Corresponds to LAMMPS cflength keyword.
Definition at line 303 of file InterfaceLammps.h.
Referenced by addNeighbor(), getForces(), getForcesChi(), getForcesDevelop(), getMaxCutoffRadius(), getMaxCutoffRadiusOverall(), initialize(), InterfaceLammps(), processDevelop(), setBoxVectors(), and setLocalAtomPositions().
◆ cfenergy
|
protected |
Corresponds to LAMMPS cfenergy keyword.
Definition at line 305 of file InterfaceLammps.h.
Referenced by getAtomicEnergy(), getEnergy(), getForces(), getForcesChi(), getForcesDevelop(), initialize(), and InterfaceLammps().
◆ emap
|
protected |
Corresponds to LAMMPS map keyword.
Definition at line 307 of file InterfaceLammps.h.
Referenced by initialize().
◆ indexMap
|
protected |
Map from LAMMPS index to n2p2 atom index.
Definition at line 309 of file InterfaceLammps.h.
Referenced by addNeighbor(), and setLocalAtoms().
◆ ignoreType
|
protected |
True if atoms of this LAMMPS type will be ignored.
Definition at line 311 of file InterfaceLammps.h.
Referenced by addNeighbor(), initialize(), and setLocalAtoms().
◆ mapTypeToElement
|
protected |
Map from LAMMPS type to n2p2 element index.
Definition at line 313 of file InterfaceLammps.h.
Referenced by addNeighbor(), initialize(), and setLocalAtoms().
◆ mapElementToType
|
protected |
Map from n2p2 element index to LAMMPS type.
Definition at line 315 of file InterfaceLammps.h.
Referenced by initialize().
◆ structure
|
protected |
Structure containing local atoms.
Definition at line 317 of file InterfaceLammps.h.
Referenced by addCharge(), addElectrostaticEnergy(), addNeighbor(), allocateNeighborlists(), finalizeNeighborList(), getAtomicEnergy(), getCharges(), getdChidxyz(), getdEdQ(), getEnergy(), getForces(), getForcesChi(), getForcesDevelop(), getMaxCutoffRadiusOverall(), getQEqParams(), initialize(), process(), processDevelop(), setBoxVectors(), setLocalAtomPositions(), setLocalAtoms(), setLocalTags(), setLocalTags(), and writeToFile().
◆ isElecDone
|
protected |
True if first NN is calculated.
Definition at line 319 of file InterfaceLammps.h.
Referenced by InterfaceLammps(), process(), and setElecDone().
The documentation for this class was generated from the following files:
- /home/runner/work/n2p2/n2p2/src/libnnpif/LAMMPS/InterfaceLammps.h
- /home/runner/work/n2p2/n2p2/src/libnnpif/LAMMPS/InterfaceLammps.cpp