Namespaces | |
| namespace | settings |
Classes | |
| struct | Atom |
| Storage for a single atom. More... | |
| class | CompactFunction |
| A general function with compact support. More... | |
| class | CoreFunction |
| class | CutoffFunction |
| class | Dataset |
| Collect and process large data sets. More... | |
| class | Element |
| Contains element-specific data. More... | |
| class | ElementCabana |
| Derived Cabana class for element-specific data. More... | |
| class | ElementMap |
| Contains element map. More... | |
| struct | ErfcBuf |
| Helper class to store previously calculated values of erfc() that are needed during the charge equilibration. More... | |
| struct | EwaldGlobalSettings |
| struct | EwaldParameters |
| class | EwaldSetup |
| Setup data for Ewald summation. More... | |
| class | EwaldStructureData |
| class | EwaldTruncJackson |
| class | EwaldTruncKolafaFixR |
| class | EwaldTruncKolafaOptEta |
| class | GradientDescent |
| Weight updates based on simple gradient descent methods. More... | |
| class | IEwaldTrunc |
| class | InterfaceLammps |
| class | KalmanFilter |
| Implementation of the Kalman filter method. More... | |
| class | KspaceGrid |
| class | Kvector |
| class | Log |
| Logging class for library output. More... | |
| class | Mode |
| Base class for all NNP applications. More... | |
| class | ModeCabana |
| Derived Cabana main NNP class. More... | |
| class | NeuralNetwork |
| This class implements a feed-forward neural network. More... | |
| class | Prediction |
| class | ScreeningFunction |
| A screening functions for use with electrostatics. More... | |
| class | SetupAnalysis |
| class | Stopwatch |
| Implements a simple stopwatch on different platforms. More... | |
| struct | Structure |
| Storage for one atomic configuration. More... | |
| class | SymFnc |
| Symmetry function base class. More... | |
| class | SymFncBaseComp |
| Symmetry function base class for SFs with compact support. More... | |
| class | SymFncBaseCompAng |
| Intermediate symmetry function class for angular SFs with compact support. More... | |
| class | SymFncBaseCompAngWeighted |
| Intermediate symmetry function class for weighted angular compact SFs. More... | |
| class | SymFncBaseCutoff |
| Intermediate class for SFs based on cutoff functions. More... | |
| class | SymFncBaseExpAng |
| Intermediate class for angular SFs based on cutoffs and exponentials. More... | |
| class | SymFncCompAngn |
| Narrow angular symmetry function with compact support (type 21) More... | |
| class | SymFncCompAngnWeighted |
| Weighted narrow angular symmetry function with compact support (type 24) More... | |
| class | SymFncCompAngw |
| Wide angular symmetry function with compact support (type 22) More... | |
| class | SymFncCompAngwWeighted |
| Weighted wide angular symmetry function with compact support (type 25) More... | |
| class | SymFncCompRad |
| Radial symmetry function with compact support (type 20) More... | |
| class | SymFncCompRadWeighted |
| Weighted radial symmetry function with compact support (type 23) More... | |
| class | SymFncExpAngn |
| Angular symmetry function (type 3) More... | |
| class | SymFncExpAngnWeighted |
| Weighted angular symmetry function (type 13) More... | |
| class | SymFncExpAngw |
| Angular symmetry function (type 9) More... | |
| class | SymFncExpRad |
| Radial symmetry function (type 2) More... | |
| class | SymFncExpRadWeighted |
| Weighted radial symmetry function (type 12) More... | |
| class | SymFncStatistics |
| class | SymGrp |
| class | SymGrpBaseComp |
| class | SymGrpBaseCompAng |
| class | SymGrpBaseCompAngWeighted |
| class | SymGrpBaseCutoff |
| class | SymGrpBaseExpAng |
| class | SymGrpCompAngn |
| Narrow angular symmetry function with compact support (type 21) More... | |
| class | SymGrpCompAngnWeighted |
| Weighted narrow angular symmetry function with compact support (type 24) More... | |
| class | SymGrpCompAngw |
| Wide angular symmetry function with compact support (type 22) More... | |
| class | SymGrpCompAngwWeighted |
| Weighted wide angular symmetry function with compact support (type 25) More... | |
| class | SymGrpCompRad |
| Radial symmetry function with compact support (type 20) More... | |
| class | SymGrpCompRadWeighted |
| Weighted radial symmetry function with compact support (type 23) More... | |
| class | SymGrpExpAngn |
| Angular symmetry function group (type 3) More... | |
| class | SymGrpExpAngnWeighted |
| Weighted angular symmetry function group (type 13) More... | |
| class | SymGrpExpAngw |
| Angular symmetry function group (type 3) More... | |
| class | SymGrpExpRad |
| Radial symmetry function group (type 2) More... | |
| class | SymGrpExpRadWeighted |
| Weighted radial symmetry function group (type 12) More... | |
| class | Training |
| Training methods. More... | |
| class | Updater |
| Base class for different weight update methods. More... | |
| struct | Vec3D |
| Vector in 3 dimensional real space. More... | |
Enumerations | |
| enum class | EWALDTruncMethod { JACKSON_CATLOW , KOLAFA_PERRAM } |
| enum class | KSPACESolver { EWALD_SUM , PPPM , EWALD_SUM_LAMMPS } |
Functions | |
| NeuralNetwork::ActivationFunction | activationFromString (std::string c) |
| Convert string to activation function. | |
| vector< string > | split (std::string const &input, char delimiter=' ') |
| Split string at each delimiter. | |
| string | trim (std::string const &line, std::string const &whitespace=" \t") |
| Remove leading and trailing whitespaces from string. | |
| string | reduce (std::string const &line, std::string const &whitespace=" \t", std::string const &fill=" ") |
| Replace multiple whitespaces with fill. | |
| string | pad (string const &input, size_t num, char fill, bool right) |
| string | strpr (const char *format,...) |
| String version of printf function. | |
| string | cap (std::string word) |
| Capitalize first letter of word. | |
| vector< string > | createFileHeader (vector< string > const &title, vector< size_t > const &colSize, vector< string > const &colName, vector< string > const &colInfo, char const &commentChar) |
| void | appendLinesToFile (std::ofstream &file, std::vector< std::string > const lines) |
| Append multiple lines of strings to open file stream. | |
| void | appendLinesToFile (FILE *const &file, std::vector< std::string > const lines) |
| Append multiple lines of strings to open file pointer. | |
| map< size_t, vector< double > > | readColumnsFromFile (string fileName, vector< size_t > columns, char comment) |
| double | pow_int (double x, int n) |
| Integer version of power function, "fast exponentiation algorithm". | |
| std::string | pad (std::string const &input, std::size_t num, char fill=' ', bool right=true) |
| Pad string to given length with fill character. | |
| std::vector< std::string > | createFileHeader (std::vector< std::string > const &title, std::vector< std::size_t > const &colLength, std::vector< std::string > const &colName, std::vector< std::string > const &colInfo, char const &commentChar='#') |
| Generate output file header with info section and column labels. | |
| std::map< std::size_t, std::vector< double > > | readColumnsFromFile (std::string fileName, std::vector< std::size_t > columns, char comment='#') |
| Read multiple columns of data from file. | |
| template<typename K, typename V> | |
| V const & | safeFind (std::map< K, V > const &stdMap, typename std::map< K, V >::key_type const &key) |
| Safely access map entry. | |
| template<typename T> | |
| bool | vectorContains (std::vector< T > const &stdVec, T value) |
| Test if vector contains specified value. | |
| template<typename T, typename U> | |
| bool | unorderedMapContainsKey (std::unordered_map< T, U > const &stdMap, T key) |
| Test if unordered map contains specified key. | |
| template<typename T> | |
| bool | comparePointerTargets (T *lhs, T *rhs) |
| Compare pointer targets. | |
| Vec3D | operator+ (Vec3D lhs, Vec3D const &rhs) |
| Overload + operator to implement vector addition. | |
| Vec3D | operator- (Vec3D lhs, Vec3D const &rhs) |
| Overload - operator to implement vector subtraction. | |
| Vec3D | operator- (Vec3D v) |
| Overload - operator to implement vector sign change. | |
| Vec3D | operator* (Vec3D v, double const a) |
| Overload * operator to implement multiplication with scalar. | |
| Vec3D | operator* (Vec3D const (&A)[3], Vec3D v) |
| Overload * operator to implement (left) multiplication with a matrix. | |
| Vec3D | operator/ (Vec3D v, double const a) |
| Overload / operator to implement division by scalar. | |
| Vec3D | operator* (double const a, Vec3D v) |
| Overload * operator to implement multiplication with scalar (scalar first). | |
Variables | |
| double constexpr | TrOverTk = 3.676 |
Enumeration Type Documentation
◆ EWALDTruncMethod
|
strong |
| Enumerator | |
|---|---|
| JACKSON_CATLOW | Method 0: Used by RuNNer (DOI: 10.1080/08927028808080944). |
| KOLAFA_PERRAM | Method 1: Optimized in n2p2 (DOI: 10.1080/08927029208049126). |
Definition at line 29 of file EwaldSetup.h.
◆ KSPACESolver
|
strong |
| Enumerator | |
|---|---|
| EWALD_SUM | Solver 0: Ewald summation. |
| PPPM | Solver 1: PPPM. |
| EWALD_SUM_LAMMPS | Solver 2: Ewald summation in LAMMPS. |
Definition at line 26 of file Kspace.h.
Function Documentation
◆ activationFromString()
| NeuralNetwork::ActivationFunction nnp::activationFromString | ( | std::string | c | ) |
Convert string to activation function.
- Parameters
-
[in] letter String representing activation function.
- Returns
- Activation corresponding to string.
Definition at line 1078 of file NeuralNetwork.cpp.
References nnp::NeuralNetwork::AF_COS, nnp::NeuralNetwork::AF_EXP, nnp::NeuralNetwork::AF_GAUSSIAN, nnp::NeuralNetwork::AF_HARMONIC, nnp::NeuralNetwork::AF_IDENTITY, nnp::NeuralNetwork::AF_LOGISTIC, nnp::NeuralNetwork::AF_RELU, nnp::NeuralNetwork::AF_REVLOGISTIC, nnp::NeuralNetwork::AF_SOFTPLUS, and nnp::NeuralNetwork::AF_TANH.
Referenced by nnp::Mode::setupNeuralNetwork().

◆ split()
| std::vector< std::string > nnp::split | ( | std::string const & | input, |
| char | delimiter = ' ' ) |
Split string at each delimiter.
- Parameters
-
[in] input Input string. [in] delimiter Delimiter character (default ' ').
- Returns
- Vector containing the string parts.
Definition at line 33 of file utility.cpp.
Referenced by nnp::SymFnc::addCacheIndex(), nnp::Element::addSymmetryFunction(), nnp::ElementCabana::addSymmetryFunction(), nnp::Training::dataSetNormalization(), nnp::Dataset::getNumStructures(), nnp::InterfaceLammps::initialize(), main(), readColumnsFromFile(), nnp::Structure::readFromFile(), nnp::Structure::readFromLines(), nnp::ElementMap::registerElements(), nnp::SymFncBaseCompAng::setParameters(), nnp::SymFncBaseCompAngWeighted::setParameters(), nnp::SymFncBaseExpAng::setParameters(), nnp::SymFncCompRad::setParameters(), nnp::SymFncCompRadWeighted::setParameters(), nnp::SymFncExpAngnWeighted::setParameters(), nnp::SymFncExpRad::setParameters(), nnp::SymFncExpRadWeighted::setParameters(), nnp::ElementCabana::setScalingType(), nnp::SymFnc::setScalingType(), nnp::Mode::setupCutoff(), nnp::Mode::setupElectrostatics(), nnp::ModeCabana< t_device >::setupElementMap(), nnp::Mode::setupElements(), nnp::ModeCabana< t_device >::setupElements(), nnp::Training::setupFileOutput(), nnp::Mode::setupNeuralNetwork(), nnp::ModeCabana< t_device >::setupNeuralNetwork(), nnp::ModeCabana< t_device >::setupNeuralNetworkWeights(), nnp::Training::setupSelectionMode(), nnp::Mode::setupSymmetryFunctionCache(), nnp::Mode::setupSymmetryFunctions(), nnp::ModeCabana< t_device >::setupSymmetryFunctions(), and nnp::Structure::writeToFilePoscar().
◆ trim()
| std::string nnp::trim | ( | std::string const & | line, |
| std::string const & | whitespace = " \t" ) |
Remove leading and trailing whitespaces from string.
- Parameters
-
[in] line Input string. [in] whitespace All characters identified as whitespace (default: " \t").
- Returns
- String without leading and trailing whitespaces.
Definition at line 47 of file utility.cpp.
Referenced by nnp::InterfaceLammps::initialize(), and reduce().
◆ reduce()
| std::string nnp::reduce | ( | std::string const & | line, |
| std::string const & | whitespace = " \t", | ||
| std::string const & | fill = " " ) |
Replace multiple whitespaces with fill.
- Parameters
-
[in] line Input string. [in] whitespace All characters identified as whitespace (default: " \t"). [in] fill Replacement character (default: " ").
- Returns
- String with fill replacements.
Calls trim() at the beginning.
Definition at line 60 of file utility.cpp.
References trim().
Referenced by nnp::Element::addSymmetryFunction(), nnp::ElementCabana::addSymmetryFunction(), nnp::Dataset::getNumStructures(), nnp::InterfaceLammps::initialize(), main(), nnp::settings::Settings::parseLines(), readColumnsFromFile(), nnp::Structure::readFromFile(), nnp::Structure::readFromLines(), nnp::ElementMap::registerElements(), nnp::SymFncBaseCompAng::setParameters(), nnp::SymFncBaseCompAngWeighted::setParameters(), nnp::SymFncBaseExpAng::setParameters(), nnp::SymFncCompRad::setParameters(), nnp::SymFncCompRadWeighted::setParameters(), nnp::SymFncExpAngnWeighted::setParameters(), nnp::SymFncExpRad::setParameters(), nnp::SymFncExpRadWeighted::setParameters(), nnp::ElementCabana::setScalingType(), nnp::SymFnc::setScalingType(), nnp::Mode::setupElectrostatics(), nnp::ModeCabana< t_device >::setupElementMap(), nnp::Mode::setupElements(), nnp::ModeCabana< t_device >::setupElements(), nnp::Training::setupFileOutput(), nnp::Mode::setupNeuralNetwork(), nnp::ModeCabana< t_device >::setupNeuralNetwork(), nnp::ModeCabana< t_device >::setupNeuralNetworkWeights(), nnp::Mode::setupSymmetryFunctions(), and nnp::ModeCabana< t_device >::setupSymmetryFunctions().
◆ pad() [1/2]
| string nnp::pad | ( | string const & | input, |
| size_t | num, | ||
| char | fill, | ||
| bool | right ) |
Definition at line 79 of file utility.cpp.
References pad().
Referenced by pad(), nnp::SymFnc::parameterInfo(), nnp::SymFncBaseComp::parameterInfo(), nnp::SymFncBaseCompAng::parameterInfo(), nnp::SymFncBaseCompAngWeighted::parameterInfo(), nnp::SymFncBaseCutoff::parameterInfo(), nnp::SymFncBaseExpAng::parameterInfo(), nnp::SymFncCompRad::parameterInfo(), nnp::SymFncExpAngnWeighted::parameterInfo(), nnp::SymFncExpRad::parameterInfo(), and nnp::SymFncExpRadWeighted::parameterInfo().

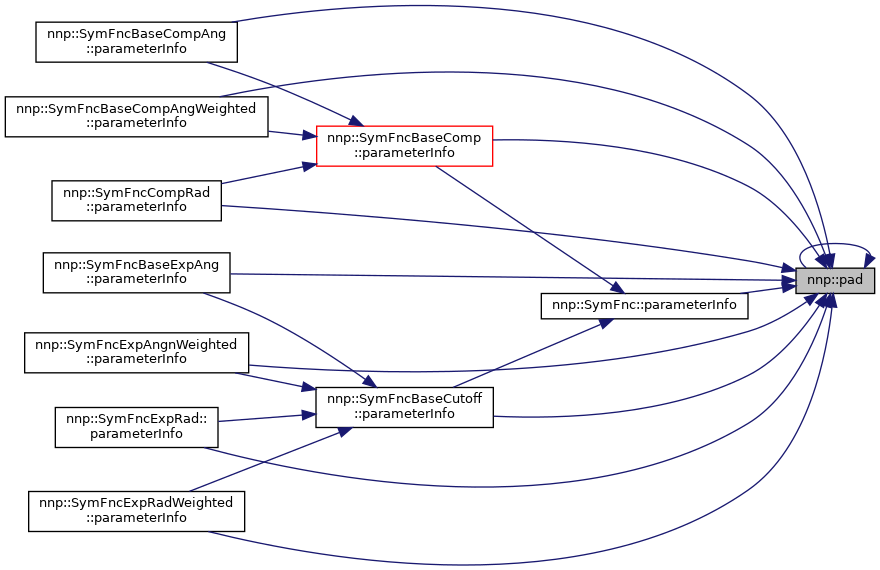
◆ strpr()
| std::string nnp::strpr | ( | const char * | format, |
| ... ) |
String version of printf function.
Definition at line 90 of file utility.cpp.
References STRPR_MAXBUF.
Referenced by nnp::SymFnc::addCacheIndex(), nnp::Training::addTrainingLogEntry(), nnp::Training::addTrainingLogEntry(), nnp::Training::addTrainingLogEntry(), nnp::Training::allocateArrays(), nnp::Training::calculateError(), nnp::Training::calculateErrorEpoch(), nnp::Training::calculateNeighborLists(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), nnp::Training::checkSelectionMode(), nnp::Dataset::collectSymmetryFunctionStatistics(), nnp::Dataset::combineFiles(), nnp::Training::dataSetNormalization(), nnp::Dataset::distributeStructures(), nnp::SymFncBaseCompAng::getCacheIdentifiers(), nnp::SymFncBaseCompAngWeighted::getCacheIdentifiers(), nnp::SymFncBaseExpAng::getCacheIdentifiers(), nnp::SymFncCompRad::getCacheIdentifiers(), nnp::SymFncCompRadWeighted::getCacheIdentifiers(), nnp::SymFncExpAngnWeighted::getCacheIdentifiers(), nnp::SymFncExpRad::getCacheIdentifiers(), nnp::SymFncExpRadWeighted::getCacheIdentifiers(), nnp::Atom::getChargeLine(), nnp::ElementMap::getElementsString(), nnp::Structure::getEnergyLine(), nnp::SymFncStatistics::getExtrapolationWarningLines(), nnp::Atom::getForcesLines(), nnp::SymFncBaseCompAng::getSettingsLine(), nnp::SymFncBaseCompAngWeighted::getSettingsLine(), nnp::SymFncBaseExpAng::getSettingsLine(), nnp::SymFncCompRad::getSettingsLine(), nnp::SymFncCompRadWeighted::getSettingsLine(), nnp::SymFncExpAngnWeighted::getSettingsLine(), nnp::SymFncExpRad::getSettingsLine(), nnp::SymFncExpRadWeighted::getSettingsLine(), nnp::Atom::info(), nnp::Atom::Neighbor::info(), nnp::CoreFunction::info(), nnp::ElementMap::info(), nnp::GradientDescent::info(), nnp::KalmanFilter::info(), nnp::NeuralNetwork::info(), nnp::ScreeningFunction::info(), nnp::Structure::info(), nnp::InterfaceLammps::initialize(), nnp::Training::initializeWeightsMemory(), nnp::Mode::loadSettingsFile(), nnp::EwaldSetup::logEwaldCutoffs(), nnp::Training::loop(), main(), nnp::SymFnc::parameterInfo(), nnp::SymFncBaseComp::parameterInfo(), nnp::SymFncBaseCompAng::parameterInfo(), nnp::SymFncBaseCompAngWeighted::parameterInfo(), nnp::SymFncBaseCutoff::parameterInfo(), nnp::SymFncBaseExpAng::parameterInfo(), nnp::SymFncCompRad::parameterInfo(), nnp::SymFncExpAngnWeighted::parameterInfo(), nnp::SymFncExpRad::parameterInfo(), nnp::SymFncExpRadWeighted::parameterInfo(), nnp::SymFncBaseCompAng::parameterLine(), nnp::SymFncBaseCompAngWeighted::parameterLine(), nnp::SymFncBaseExpAng::parameterLine(), nnp::SymFncCompRad::parameterLine(), nnp::SymFncCompRadWeighted::parameterLine(), nnp::SymFncExpAngnWeighted::parameterLine(), nnp::SymFncExpRad::parameterLine(), nnp::SymFncExpRadWeighted::parameterLine(), nnp::SymGrpBaseCompAng::parameterLines(), nnp::SymGrpBaseCompAngWeighted::parameterLines(), nnp::SymGrpBaseExpAng::parameterLines(), nnp::SymGrpCompRad::parameterLines(), nnp::SymGrpCompRadWeighted::parameterLines(), nnp::SymGrpExpAngnWeighted::parameterLines(), nnp::SymGrpExpRad::parameterLines(), nnp::SymGrpExpRadWeighted::parameterLines(), nnp::settings::Settings::parseLines(), nnp::Dataset::prepareNumericForces(), nnp::Training::printEpoch(), nnp::Training::printHeader(), nnp::Training::randomizeNeuralNetworkWeights(), nnp::settings::Settings::readFile(), nnp::Mode::readNeuralNetworkWeights(), runTest(), runTest(), nnp::settings::Settings::sanityCheck(), nnp::ElementCabana::scalingLine(), nnp::SymFnc::scalingLine(), nnp::Training::selectSets(), nnp::SymFncBaseComp::setCompactFunction(), nnp::SymFncBaseCutoff::setCutoffFunction(), nnp::Element::setScalingNone(), nnp::Training::setStage(), nnp::Mode::setupCutoff(), nnp::Mode::setupElectrostatics(), nnp::Mode::setupElementMap(), nnp::ModeCabana< t_device >::setupElementMap(), nnp::Mode::setupElements(), nnp::ModeCabana< t_device >::setupElements(), nnp::Training::setupFileOutput(), nnp::Dataset::setupMPI(), nnp::Mode::setupNeuralNetwork(), nnp::ModeCabana< t_device >::setupNeuralNetwork(), nnp::Mode::setupNeuralNetworkWeights(), nnp::ModeCabana< t_device >::setupNeuralNetworkWeights(), nnp::Mode::setupNormalization(), nnp::Dataset::setupRandomNumberGenerator(), nnp::Training::setupSelectionMode(), nnp::Mode::setupSymmetryFunctionCache(), nnp::Mode::setupSymmetryFunctionGroups(), nnp::ModeCabana< t_device >::setupSymmetryFunctionGroups(), nnp::Mode::setupSymmetryFunctionMemory(), nnp::Mode::setupSymmetryFunctions(), nnp::ModeCabana< t_device >::setupSymmetryFunctions(), nnp::Mode::setupSymmetryFunctionScaling(), nnp::ModeCabana< t_device >::setupSymmetryFunctionScaling(), nnp::Mode::setupSymmetryFunctionStatistics(), nnp::Training::setupTraining(), nnp::Training::setupUpdatePlan(), nnp::GradientDescent::status(), nnp::KalmanFilter::status(), nnp::ElementMap::symbolFromAtomicNumber(), nnp::Element::updateSymmetryFunctionStatistics(), nnp::Dataset::writeAtomicEnvironmentFile(), nnp::NeuralNetwork::writeConnections(), nnp::Training::writeHardness(), nnp::Training::writeHardnessEpoch(), nnp::Training::writeLearningCurve(), nnp::Dataset::writeNeighborHistogram(), nnp::Dataset::writeNeighborLists(), nnp::Training::writeNeuronStatistics(), nnp::Training::writeNeuronStatisticsEpoch(), nnp::Training::writeSetsToFiles(), nnp::Dataset::writeSymmetryFunctionFile(), nnp::Dataset::writeSymmetryFunctionHistograms(), nnp::Dataset::writeSymmetryFunctionScaling(), nnp::SetupAnalysis::writeSymmetryFunctionShape(), nnp::Training::writeTimingData(), nnp::Structure::writeToFile(), nnp::Structure::writeToFilePoscar(), nnp::Structure::writeToFileXyz(), nnp::Training::writeUpdaterStatus(), nnp::Training::writeWeights(), and nnp::Training::writeWeightsEpoch().
◆ cap()
| std::string nnp::cap | ( | std::string | word | ) |
Capitalize first letter of word.
- Parameters
-
[in] word String to capitalize.
- Returns
- Input string with first letter in capital letters.
Definition at line 104 of file utility.cpp.
Referenced by nnp::Mode::setupNeuralNetworkWeights().

◆ createFileHeader() [1/2]
| vector< string > nnp::createFileHeader | ( | vector< string > const & | title, |
| vector< size_t > const & | colSize, | ||
| vector< string > const & | colName, | ||
| vector< string > const & | colInfo, | ||
| char const & | commentChar ) |
Definition at line 111 of file utility.cpp.
Referenced by nnp::Training::calculateError(), nnp::Training::dataSetNormalization(), main(), nnp::Training::setupTraining(), nnp::GradientDescent::statusHeader(), nnp::KalmanFilter::statusHeader(), nnp::NeuralNetwork::writeConnections(), nnp::Training::writeLearningCurve(), nnp::Dataset::writeNeighborHistogram(), nnp::Training::writeNeuronStatistics(), nnp::Dataset::writeSymmetryFunctionHistograms(), nnp::Dataset::writeSymmetryFunctionScaling(), nnp::SetupAnalysis::writeSymmetryFunctionShape(), and nnp::Training::writeTimingData().
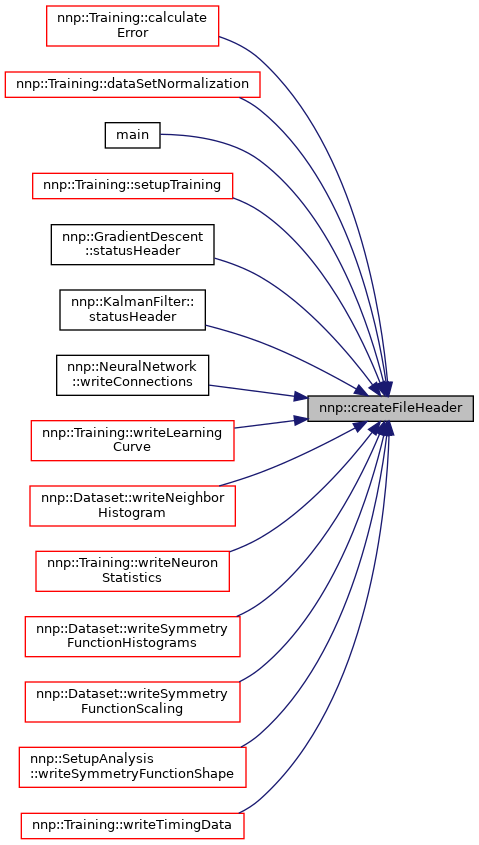
◆ appendLinesToFile() [1/2]
| void nnp::appendLinesToFile | ( | std::ofstream & | file, |
| std::vector< std::string > const | lines ) |
Append multiple lines of strings to open file stream.
- Parameters
-
[in,out] file File stream to append lines to. [in] lines Vector of strings with line content.
Definition at line 225 of file utility.cpp.
Referenced by nnp::Training::calculateError(), nnp::Training::dataSetNormalization(), main(), nnp::Training::setupTraining(), nnp::NeuralNetwork::writeConnections(), nnp::Training::writeLearningCurve(), nnp::Dataset::writeNeighborHistogram(), nnp::Training::writeNeuronStatistics(), nnp::Dataset::writeSymmetryFunctionHistograms(), nnp::Dataset::writeSymmetryFunctionScaling(), nnp::SetupAnalysis::writeSymmetryFunctionShape(), nnp::Training::writeTimingData(), and nnp::Training::writeUpdaterStatus().
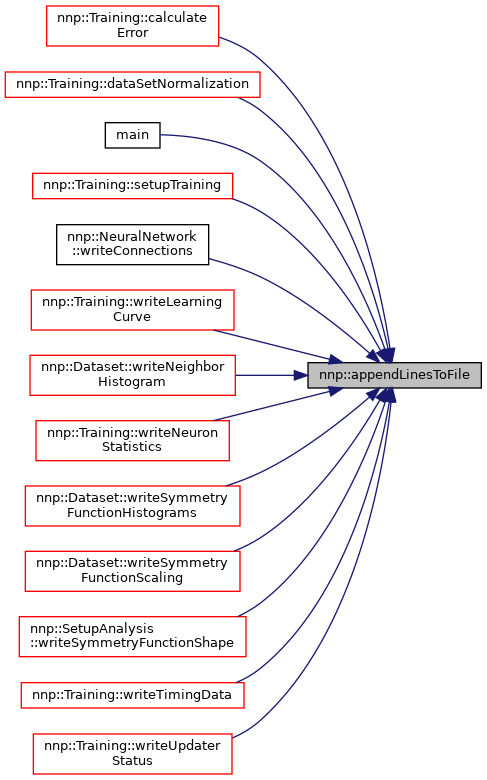
◆ appendLinesToFile() [2/2]
| void nnp::appendLinesToFile | ( | FILE *const & | file, |
| std::vector< std::string > const | lines ) |
Append multiple lines of strings to open file pointer.
- Parameters
-
[in,out] file File pointer. [in] lines Vector of strings with line content.
Definition at line 236 of file utility.cpp.
◆ readColumnsFromFile() [1/2]
| map< size_t, vector< double > > nnp::readColumnsFromFile | ( | string | fileName, |
| vector< size_t > | columns, | ||
| char | comment ) |
Definition at line 247 of file utility.cpp.
References reduce(), and split().
Referenced by nnp::Mode::readNeuralNetworkWeights(), and nnp::Mode::setupElectrostatics().
◆ pow_int()
| double nnp::pow_int | ( | double | x, |
| int | n ) |
Integer version of power function, "fast exponentiation algorithm".
- Parameters
-
[in] x Base number. [in] n Integer exponent.
- Returns
- x^n
Definition at line 292 of file utility.cpp.
Referenced by nnp::SymFncExpAngn::calculate(), nnp::SymFncExpAngnWeighted::calculate(), nnp::SymFncExpAngw::calculate(), nnp::SymGrpExpAngn::calculate(), nnp::SymGrpExpAngnWeighted::calculate(), and nnp::SymGrpExpAngw::calculate().
◆ pad() [2/2]
| std::string nnp::pad | ( | std::string const & | input, |
| std::size_t | num, | ||
| char | fill = ' ', | ||
| bool | right = true ) |
Pad string to given length with fill character.
- Parameters
-
[in] input Input string. [in] num Desired width of padded string. [in] fill Padding character. [in] right If true, pad right. Pad left otherwise.
- Returns
- String padded to desired length or original string if length of old string is not shorter than desired length.
◆ createFileHeader() [2/2]
| std::vector< std::string > nnp::createFileHeader | ( | std::vector< std::string > const & | title, |
| std::vector< std::size_t > const & | colLength, | ||
| std::vector< std::string > const & | colName, | ||
| std::vector< std::string > const & | colInfo, | ||
| char const & | commentChar = '#' ) |
Generate output file header with info section and column labels.
- Parameters
-
[in] title Multiple lines of title text. [in] colLength Length of the individual columns. [in] colName Names of the individual columns. [in] colInfo Description of the individual columns. [in] commentChar Character used for starting each line and to create separator lines.
- Returns
- Vector of strings, one entry per line.
◆ readColumnsFromFile() [2/2]
| std::map< std::size_t, std::vector< double > > nnp::readColumnsFromFile | ( | std::string | fileName, |
| std::vector< std::size_t > | columns, | ||
| char | comment = '#' ) |
Read multiple columns of data from file.
- Parameters
-
[in] fileName Name of data file. [in] columns Vector with column indices of interest (starting with 0). [in] comment Lines starting with comment sign are ignored.
- Returns
- Map from column index to vector of data.
◆ safeFind()
| V const & nnp::safeFind | ( | std::map< K, V > const & | stdMap, |
| typename std::map< K, V >::key_type const & | key ) |
Safely access map entry.
- Parameters
-
[in] stdMap Map to search in. [in] key Key requested.
The .at() member function of maps is available only for C++11.
Definition at line 144 of file utility.h.
Referenced by nnp::SymFnc::getPrintFormat(), nnp::SymGrp::getPrintFormatCommon(), nnp::SymGrp::getPrintFormatMember(), nnp::ElementMap::index(), nnp::ElementMap::symbol(), and nnp::Dataset::writeSymmetryFunctionHistograms().
◆ vectorContains()
| bool nnp::vectorContains | ( | std::vector< T > const & | stdVec, |
| T | value ) |
Test if vector contains specified value.
- Parameters
-
[in] stdVec Vector to search in. [in] value Value to search for.
- Returns
Trueif value is located in vector.
Definition at line 166 of file utility.h.
Referenced by nnp::Element::getCutoffRadii(), nnp::Mode::setupCutoffMatrix(), and nnp::Structure::setupNeighborCutoffMap().
◆ unorderedMapContainsKey()
| bool nnp::unorderedMapContainsKey | ( | std::unordered_map< T, U > const & | stdMap, |
| T | key ) |
Test if unordered map contains specified key.
- Parameters
-
[in] stdMap Map to search in. [in] key Key to search for.
- Returns
Trueif key is located in map.
Definition at line 179 of file utility.h.
Referenced by nnp::Atom::calculateNumNeighbors(), and nnp::Atom::getStoredMinNumNeighbors().
◆ comparePointerTargets()
| bool nnp::comparePointerTargets | ( | T * | lhs, |
| T * | rhs ) |
Compare pointer targets.
- Parameters
-
[in] lhs Pointer to left side of comparison. [in] rhs Pointer to right side of comparison.
- Returns
Trueif dereferencedlhs< dereferencedrhs,Falseotherwise.
Definition at line 194 of file utility.h.
Referenced by nnp::Element::setupSymmetryFunctionGroups(), nnp::SymGrpCompAngn::sortMembers(), nnp::SymGrpCompAngnWeighted::sortMembers(), nnp::SymGrpCompAngw::sortMembers(), nnp::SymGrpCompAngwWeighted::sortMembers(), nnp::SymGrpCompRad::sortMembers(), nnp::SymGrpCompRadWeighted::sortMembers(), nnp::SymGrpExpAngn::sortMembers(), nnp::SymGrpExpAngnWeighted::sortMembers(), nnp::SymGrpExpAngw::sortMembers(), nnp::SymGrpExpRad::sortMembers(), nnp::SymGrpExpRadWeighted::sortMembers(), and nnp::Element::sortSymmetryFunctions().
◆ operator+()
◆ operator-() [1/2]
◆ operator-() [2/2]
◆ operator*() [1/3]
◆ operator*() [2/3]
◆ operator/()
◆ operator*() [3/3]
Variable Documentation
◆ TrOverTk
|
constexpr |
Definition at line 14 of file EwaldTruncKolafaOptEta.cpp.
Referenced by nnp::EwaldTruncKolafaOptEta::calculateEta().