#include <Training.h>
Classes | |
| struct | Property |
| Specific training quantity (e.g. energies, forces, charges). More... | |
| struct | PropertyMap |
| Map of all training properties. More... | |
| struct | SubCandidate |
| Contains update candidate which is grouped with others to specific parent update candidate (e.g. More... | |
| struct | UpdateCandidate |
| Contains location of one update candidate (energy or force). More... | |
Public Types | |
| enum | UpdaterType { UT_GD , UT_KF , UT_LM } |
| Type of update routine. More... | |
| enum | ParallelMode { PM_TRAIN_RK0 , PM_TRAIN_ALL } |
| Training parallelization mode. More... | |
| enum | JacobianMode { JM_SUM , JM_TASK , JM_FULL } |
| Jacobian matrix preparation mode. More... | |
| enum | UpdateStrategy { US_COMBINED , US_ELEMENT } |
| Update strategies available for Training. More... | |
| enum | SelectionMode { SM_RANDOM , SM_SORT , SM_THRESHOLD } |
| How update candidates are selected during Training. More... | |
 Public Types inherited from nnp::Mode Public Types inherited from nnp::Mode | |
| enum class | NNPType { HDNNP_2G = 2 , HDNNP_4G = 4 , HDNNP_Q = 10 } |
Public Member Functions | |
| Training () | |
| Constructor. | |
| ~Training () | |
| Destructor, updater vector needs to be cleaned. | |
| void | selectSets () |
| Randomly select training and test set structures. | |
| void | writeSetsToFiles () |
| Write training and test set to separate files (train.data and test.data, same format as input.data). | |
| void | initializeWeights () |
| Initialize weights for all elements. | |
| void | initializeWeightsMemory (UpdateStrategy updateStrategy=US_COMBINED) |
| Initialize weights vector according to update strategy. | |
| void | setStage (std::size_t stage) |
| Set training stage (if multiple stages are needed for NNP type). | |
| void | dataSetNormalization () |
| Apply normalization based on initial weights prediction. | |
| void | setupTraining () |
| General training settings and setup of weight update routine. | |
| std::vector< std::string > | setupNumericDerivCheck () |
| Set up numeric weight derivatives check. | |
| void | calculateNeighborLists () |
| Calculate neighbor lists for all structures. | |
| void | calculateError (std::map< std::string, std::pair< std::string, std::string > > const fileNames) |
| Calculate error metrics for all structures. | |
| void | calculateErrorEpoch () |
| Calculate error metrics per epoch for all structures with file names used in training loop. | |
| void | calculateChargeErrorVec (Structure const &s, Eigen::VectorXd &cVec, double &cNorm) |
| Calculate vector of charge errors in one structure. | |
| void | printHeader () |
| Print training loop header on screen. | |
| void | printEpoch () |
| Print preferred error metric and timing information on screen. | |
| void | writeWeights (std::string const &nnName, std::string const &fileNameFormat) const |
| Write weights to files (one file for each element). | |
| void | writeWeightsEpoch () const |
| Write weights to files during training loop. | |
| void | writeHardness (std::string const &fileNameFormat) const |
| Write hardness to files (one file for each element). | |
| void | writeHardnessEpoch () const |
| Write hardness to files during training loop. | |
| void | writeLearningCurve (bool append, std::string const fileName="learning-curve.out") const |
| Write current RMSEs and epoch information to file. | |
| void | writeNeuronStatistics (std::string const &nnName, std::string const &fileName) const |
| Write neuron statistics collected since last invocation. | |
| void | writeNeuronStatisticsEpoch () const |
| Write neuron statistics during training loop. | |
| void | resetNeuronStatistics () |
| Reset neuron statistics for all elements. | |
| void | writeUpdaterStatus (bool append, std::string const fileNameFormat="updater.%03zu.out") const |
| Write updater information to file. | |
| void | sortUpdateCandidates (std::string const &property) |
| Sort update candidates with descending RMSE. | |
| void | shuffleUpdateCandidates (std::string const &property) |
| Shuffle update candidates. | |
| void | checkSelectionMode () |
| Check if selection mode should be changed. | |
| void | loop () |
| Execute main training loop. | |
| void | setEpochSchedule () |
| Select energies/forces schedule for one epoch. | |
| void | update (std::string const &property) |
| Perform one update. | |
| double | getSingleWeight (std::size_t element, std::size_t index) |
| Get a single weight value. | |
| void | setSingleWeight (std::size_t element, std::size_t index, double value) |
| Set a single weight value. | |
| std::vector< std::vector< double > > | calculateWeightDerivatives (Structure *structure) |
| Calculate derivatives of energy with respect to weights. | |
| std::vector< std::vector< double > > | calculateWeightDerivatives (Structure *structure, std::size_t atom, std::size_t component) |
| Calculate derivatives of force with respect to weights. | |
| void | setTrainingLogFileName (std::string fileName) |
| Set training log file name. | |
| std::size_t | getNumConnections (std::string id="short") const |
| Get total number of NN connections. | |
| std::vector< std::size_t > | getNumConnectionsPerElement (std::string id="short") const |
| Get number of NN connections for each element. | |
| std::vector< std::size_t > | getConnectionOffsets (std::string id="short") const |
| Get offsets of NN connections for each element. | |
| void | dPdc (std::string property, Structure &structure, std::vector< std::vector< double > > &dEdc) |
| Compute derivatives of property with respect to weights. | |
| void | dPdcN (std::string property, Structure &structure, std::vector< std::vector< double > > &dEdc, double delta=1.0E-4) |
| Compute numeric derivatives of property with respect to weights. | |
 Public Member Functions inherited from nnp::Dataset Public Member Functions inherited from nnp::Dataset | |
| Dataset () | |
| Constructor, initialize members. | |
| ~Dataset () | |
| Destructor. | |
| void | setupMPI () |
| Initialize MPI with MPI_COMM_WORLD. | |
| void | setupMPI (MPI_Comm *communicator) |
| Initialize MPI with given communicator. | |
| void | setupRandomNumberGenerator () |
| Initialize random number generator. | |
| std::size_t | getNumStructures (std::ifstream &dataFile) |
| Get number of structures in data file. | |
| int | calculateBufferSize (Structure const &structure) const |
| Calculate buffer size required to communicate structure via MPI. | |
| int | sendStructure (Structure const &structure, int dest) const |
| Send one structure to destination process. | |
| int | recvStructure (Structure *structure, int src) |
| Receive one structure from source process. | |
| int | distributeStructures (bool randomize, bool excludeRank0=false, std::string const &fileName="input.data") |
| Read data file and distribute structures among processors. | |
| std::size_t | prepareNumericForces (Structure &original, double delta) |
| Prepare numeric force check for a single structure. | |
| void | toNormalizedUnits () |
| Switch all structures to normalized units. | |
| void | toPhysicalUnits () |
| Switch all structures to physical units. | |
| void | collectSymmetryFunctionStatistics () |
| Collect symmetry function statistics from all processors. | |
| void | writeSymmetryFunctionScaling (std::string const &fileName="scaling.data") |
| Write symmetry function scaling values to file. | |
| void | writeSymmetryFunctionHistograms (std::size_t numBins, std::string fileNameFormat="sf.%03zu.%04zu.histo") |
| Calculate and write symmetry function histograms. | |
| void | writeSymmetryFunctionFile (std::string fileName="function.data") |
| Write symmetry function legacy file ("function.data"). | |
| std::size_t | writeNeighborHistogram (std::string const &fileNameHisto="neighbors.histo", std::string const &fileNameStructure="neighbors.out") |
| Calculate and write neighbor histogram and per-structure statistics. | |
| void | sortNeighborLists () |
| Sort all neighbor lists according to element and distance. | |
| void | writeNeighborLists (std::string const &fileName="neighbor-list.data") |
| Write neighbor list file. | |
| void | writeAtomicEnvironmentFile (std::vector< std::vector< std::size_t > > neighCutoff, bool derivatives, std::string const &fileNamePrefix="atomic-env") |
| Write atomic environment file. | |
| void | collectError (std::string const &property, std::map< std::string, double > &error, std::size_t &count) const |
| Collect error metrics of a property over all MPI procs. | |
| void | combineFiles (std::string filePrefix) const |
| Combine individual MPI proc files to one. | |
 Public Member Functions inherited from nnp::Mode Public Member Functions inherited from nnp::Mode | |
| Mode () | |
| void | initialize () |
| Write welcome message with version information. | |
| void | loadSettingsFile (std::string const &fileName="input.nn") |
| Open settings file and load all keywords into memory. | |
| void | setupGeneric (std::string const &nnpDir="", bool skipNormalize=false, bool initialHardness=false) |
| Combine multiple setup routines and provide a basic NNP setup. | |
| void | setupNormalization (bool standalone=true) |
| Set up normalization. | |
| virtual void | setupElementMap () |
| Set up the element map. | |
| virtual void | setupElements () |
| Set up all Element instances. | |
| void | setupCutoff () |
| Set up cutoff function for all symmetry functions. | |
| virtual void | setupSymmetryFunctions () |
| Set up all symmetry functions. | |
| void | setupSymmetryFunctionScalingNone () |
| Set up "empy" symmetry function scaling. | |
| virtual void | setupSymmetryFunctionScaling (std::string const &fileName="scaling.data") |
| Set up symmetry function scaling from file. | |
| virtual void | setupSymmetryFunctionGroups () |
| Set up symmetry function groups. | |
| virtual void | setupSymmetryFunctionCache (bool verbose=false) |
| Set up symmetry function cache. | |
| void | setupSymmetryFunctionMemory (bool verbose=false) |
| Extract required memory dimensions for symmetry function derivatives. | |
| void | setupSymmetryFunctionStatistics (bool collectStatistics, bool collectExtrapolationWarnings, bool writeExtrapolationWarnings, bool stopOnExtrapolationWarnings) |
| Set up symmetry function statistics collection. | |
| void | setupCutoffMatrix () |
| Setup matrix storing all symmetry function cut-offs for each element. | |
| virtual void | setupNeuralNetwork () |
| Set up neural networks for all elements. | |
| virtual void | setupNeuralNetworkWeights (std::map< std::string, std::string > fileNameFormats=std::map< std::string, std::string >()) |
| Set up neural network weights from files with given name format. | |
| virtual void | setupNeuralNetworkWeights (std::string directoryPrefix, std::map< std::string, std::string > fileNameFormats=std::map< std::string, std::string >()) |
| Set up neural network weights from files with given name format. | |
| virtual void | setupElectrostatics (bool initialHardness=false, std::string directoryPrefix="", std::string fileNameFormat="hardness.%03zu.data") |
| Set up electrostatics related stuff (hardness, screening, ...). | |
| void | calculateSymmetryFunctions (Structure &structure, bool const derivatives) |
| Calculate all symmetry functions for all atoms in given structure. | |
| void | calculateSymmetryFunctionGroups (Structure &structure, bool const derivatives) |
| Calculate all symmetry function groups for all atoms in given structure. | |
| void | calculateAtomicNeuralNetworks (Structure &structure, bool const derivatives, std::string id="") |
| Calculate a single atomic neural network for a given atom and nn type. | |
| void | chargeEquilibration (Structure &structure, bool const derivativesElec) |
| Perform global charge equilibration method. | |
| void | calculateEnergy (Structure &structure) const |
| Calculate potential energy for a given structure. | |
| void | calculateCharge (Structure &structure) const |
| Calculate total charge for a given structure. | |
| void | calculateForces (Structure &structure) const |
| Calculate forces for all atoms in given structure. | |
| void | evaluateNNP (Structure &structure, bool useForces=true, bool useDEdG=true) |
| Evaluate neural network potential (includes total energy, optionally forces and in some cases charges. | |
| void | addEnergyOffset (Structure &structure, bool ref=true) |
| Add atomic energy offsets to reference energy. | |
| void | removeEnergyOffset (Structure &structure, bool ref=true) |
| Remove atomic energy offsets from reference energy. | |
| double | getEnergyOffset (Structure const &structure) const |
| Get atomic energy offset for given structure. | |
| double | getEnergyWithOffset (Structure const &structure, bool ref=true) const |
| Add atomic energy offsets and return energy. | |
| double | normalized (std::string const &property, double value) const |
| Apply normalization to given property. | |
| double | normalizedEnergy (Structure const &structure, bool ref=true) const |
| Apply normalization to given energy of structure. | |
| double | physical (std::string const &property, double value) const |
| Undo normalization for a given property. | |
| double | physicalEnergy (Structure const &structure, bool ref=true) const |
| Undo normalization for a given energy of structure. | |
| void | convertToNormalizedUnits (Structure &structure) const |
| Convert one structure to normalized units. | |
| void | convertToPhysicalUnits (Structure &structure) const |
| Convert one structure to physical units. | |
| void | logEwaldCutoffs () |
| Logs Ewald params whenever they change. | |
| std::size_t | getNumExtrapolationWarnings () const |
| Count total number of extrapolation warnings encountered for all elements and symmetry functions. | |
| void | resetExtrapolationWarnings () |
| Erase all extrapolation warnings and reset counters. | |
| NNPType | getNnpType () const |
| Getter for Mode::nnpType. | |
| double | getMeanEnergy () const |
| Getter for Mode::meanEnergy. | |
| double | getConvEnergy () const |
| Getter for Mode::convEnergy. | |
| double | getConvLength () const |
| Getter for Mode::convLength. | |
| double | getConvCharge () const |
| Getter for Mode::convCharge. | |
| double | getEwaldPrecision () const |
| Getter for Mode::ewaldSetup.precision. | |
| double | getEwaldMaxCharge () const |
| Getter for Mode::ewaldSetup.maxCharge. | |
| double | getEwaldMaxSigma () const |
| Getter for Mode::ewaldSetup.maxQsigma. | |
| EWALDTruncMethod | getEwaldTruncationMethod () const |
| Getter for Mode::ewaldSetup.truncMethod. | |
| KSPACESolver | kspaceSolver () const |
| Getter for Mode::kspaceSolver. | |
| double | getMaxCutoffRadius () const |
| Getter for Mode::maxCutoffRadius. | |
| std::size_t | getNumElements () const |
| Getter for Mode::numElements. | |
| ScreeningFunction | getScreeningFunction () const |
| Getter for Mode::screeningFunction. | |
| std::vector< std::size_t > | getNumSymmetryFunctions () const |
| Get number of symmetry functions per element. | |
| bool | useNormalization () const |
| Check if normalization is enabled. | |
| bool | settingsKeywordExists (std::string const &keyword) const |
| Check if keyword was found in settings file. | |
| std::string | settingsGetValue (std::string const &keyword) const |
| Get value for given keyword in Settings instance. | |
| std::vector< std::size_t > | pruneSymmetryFunctionsRange (double threshold) |
| Prune symmetry functions according to their range and write settings file. | |
| std::vector< std::size_t > | pruneSymmetryFunctionsSensitivity (double threshold, std::vector< std::vector< double > > sensitivity) |
| Prune symmetry functions with sensitivity analysis data. | |
| void | writePrunedSettingsFile (std::vector< std::size_t > prune, std::string fileName="output.nn") const |
| Copy settings file but comment out lines provided. | |
| void | writeSettingsFile (std::ofstream *const &file) const |
| Write complete settings file. | |
Private Member Functions | |
| bool | advance () const |
| Check if training loop should be continued. | |
| void | getWeights () |
| Get weights from neural network class. | |
| void | setWeights () |
| Set weights in neural network class. | |
| void | addTrainingLogEntry (int proc, std::size_t il, double f, std::size_t isg, std::size_t is) |
| Write energy update data to training log file. | |
| void | addTrainingLogEntry (int proc, std::size_t il, double f, std::size_t isg, std::size_t is, std::size_t ia, std::size_t ic) |
| Write force update data to training log file. | |
| void | addTrainingLogEntry (int proc, std::size_t il, double f, std::size_t isg, std::size_t is, std::size_t ia) |
| Write charge update data to training log file. | |
| void | collectDGdxia (Atom const &atom, std::size_t indexAtom, std::size_t indexComponent) |
| Collect derivative of symmetry functions with respect to one atom's coordinate. | |
| void | randomizeNeuralNetworkWeights (std::string const &id) |
| Randomly initialize specificy neural network weights. | |
| void | setupSelectionMode (std::string const &property) |
| Set selection mode for specific training property. | |
| void | setupFileOutput (std::string const &type) |
| Set file output intervals for properties and other quantities. | |
| void | setupUpdatePlan (std::string const &property) |
| Set up how often properties are updated. | |
| void | allocateArrays (std::string const &property) |
| Allocate error and Jacobian arrays for given property. | |
| void | writeTimingData (bool append, std::string const fileName="timing.out") |
| Write timing data for all clocks. | |
Private Attributes | |
| UpdaterType | updaterType |
| Updater type used. | |
| ParallelMode | parallelMode |
| Parallelization mode used. | |
| JacobianMode | jacobianMode |
| Jacobian mode used. | |
| UpdateStrategy | updateStrategy |
| Update strategy used. | |
| bool | hasUpdaters |
| If this rank performs weight updates. | |
| bool | hasStructures |
| If this rank holds structure information. | |
| bool | useForces |
| Use forces for training. | |
| bool | repeatedEnergyUpdates |
| After force update perform energy update for corresponding structure. | |
| bool | freeMemory |
| Free symmetry function memory after calculation. | |
| bool | writeTrainingLog |
| Whether training log file is written. | |
| std::size_t | stage |
| Training stage. | |
| std::size_t | numUpdaters |
| Number of updaters (depends on update strategy). | |
| std::size_t | numEpochs |
| Number of epochs requested. | |
| std::size_t | epoch |
| Current epoch. | |
| std::size_t | writeWeightsEvery |
| Write weights every this many epochs. | |
| std::size_t | writeWeightsAlways |
| Up to this epoch weights are written every epoch. | |
| std::size_t | writeNeuronStatisticsEvery |
| Write neuron statistics every this many epochs. | |
| std::size_t | writeNeuronStatisticsAlways |
| Up to this epoch neuron statistics are written every epoch. | |
| std::size_t | countUpdates |
| Update counter (for all training quantities together). | |
| std::size_t | numWeights |
| Total number of weights. | |
| double | forceWeight |
| Force update weight. | |
| std::string | trainingLogFileName |
| File name for training log. | |
| std::string | nnId |
| ID of neural network the training is working on. | |
| std::ofstream | trainingLog |
| Training log file. | |
| std::vector< int > | epochSchedule |
| Update schedule epoch (stage 1: 0 = charge update; stage 2: 0 = energy update, 1 = force update (optional)). | |
| std::vector< std::size_t > | numWeightsPerUpdater |
| Number of weights per updater. | |
| std::vector< std::size_t > | weightsOffset |
| Offset of each element's weights in combined array. | |
| std::vector< std::string > | pk |
| Vector of actually used training properties. | |
| std::vector< double > | dGdxia |
| Derivative of symmetry functions with respect to one specific atom coordinate. | |
| std::vector< std::vector< double > > | weights |
| Neural network weights and biases for each element. | |
| std::vector< Updater * > | updaters |
| Weight updater (combined or for each element). | |
| std::map< std::string, Stopwatch > | sw |
| Stopwatches for timing overview. | |
| std::mt19937_64 | rngNew |
| Per-task random number generator. | |
| std::mt19937_64 | rngGlobalNew |
| Global random number generator. | |
| PropertyMap | p |
| Actual training properties. | |
Additional Inherited Members | |
 Public Attributes inherited from nnp::Dataset Public Attributes inherited from nnp::Dataset | |
| std::vector< Structure > | structures |
| All structures in this dataset. | |
 Public Attributes inherited from nnp::Mode Public Attributes inherited from nnp::Mode | |
| ElementMap | elementMap |
| Global element map, populated by setupElementMap(). | |
| Log | log |
| Global log file. | |
 Protected Member Functions inherited from nnp::Mode Protected Member Functions inherited from nnp::Mode | |
| void | readNeuralNetworkWeights (std::string const &id, std::string const &fileName) |
| Read in weights for a specific type of neural network. | |
 Protected Attributes inherited from nnp::Dataset Protected Attributes inherited from nnp::Dataset | |
| int | myRank |
| My process ID. | |
| int | numProcs |
| Total number of MPI processors. | |
| std::size_t | numStructures |
| Total number of structures in dataset. | |
| std::string | myName |
| My processor name. | |
| MPI_Comm | comm |
| Global MPI communicator. | |
| gsl_rng * | rng |
| GSL random number generator (different seed for each MPI process). | |
| gsl_rng * | rngGlobal |
| Global GSL random number generator (equal seed for each MPI process). | |
 Protected Attributes inherited from nnp::Mode Protected Attributes inherited from nnp::Mode | |
| NNPType | nnpType |
| bool | normalize |
| bool | checkExtrapolationWarnings |
| std::size_t | numElements |
| std::vector< std::size_t > | minNeighbors |
| std::vector< double > | minCutoffRadius |
| double | maxCutoffRadius |
| double | cutoffAlpha |
| double | meanEnergy |
| double | convEnergy |
| double | convLength |
| double | convCharge |
| double | fourPiEps |
| EwaldSetup | ewaldSetup |
| KspaceGrid | kspaceGrid |
| settings::Settings | settings |
| SymFnc::ScalingType | scalingType |
| CutoffFunction::CutoffType | cutoffType |
| ScreeningFunction | screeningFunction |
| std::vector< Element > | elements |
| std::vector< std::string > | nnk |
| std::map< std::string, NNSetup > | nns |
| std::vector< std::vector< double > > | cutoffs |
| Matrix storing all symmetry function cut-offs for all elements. | |
| ErfcBuf | erfcBuf |
Detailed Description
Training methods.
Definition at line 35 of file Training.h.
Member Enumeration Documentation
◆ UpdaterType
Type of update routine.
| Enumerator | |
|---|---|
| UT_GD | Simple gradient descent methods. |
| UT_KF | Kalman filter-based methods. |
| UT_LM | Levenberg-Marquardt algorithm. |
Definition at line 39 of file Training.h.
◆ ParallelMode
Training parallelization mode.
This mode determines if and how individual MPI tasks contribute to parallel training. Note that in all cases the data set gets distributed among the MPI processes and RMSE computation is always parallelized.
Definition at line 55 of file Training.h.
◆ JacobianMode
Jacobian matrix preparation mode.
| Enumerator | |
|---|---|
| JM_SUM | No Jacobian, sum up contributions from update candidates. |
| JM_TASK | Prepare one Jacobian entry for each task, sum up within tasks. |
| JM_FULL | Prepare full Jacobian matrix. |
Definition at line 84 of file Training.h.
◆ UpdateStrategy
Update strategies available for Training.
| Enumerator | |
|---|---|
| US_COMBINED | One combined updater for all elements. |
| US_ELEMENT | Separate updaters for individual elements. |
Definition at line 95 of file Training.h.
◆ SelectionMode
How update candidates are selected during Training.
| Enumerator | |
|---|---|
| SM_RANDOM | Select candidates randomly. |
| SM_SORT | Sort candidates according to their RMSE and pick worst first. |
| SM_THRESHOLD | Select candidates randomly with RMSE above threshold. |
Definition at line 104 of file Training.h.
Constructor & Destructor Documentation
◆ Training()
| Training::Training | ( | ) |
Constructor.
Definition at line 38 of file Training.cpp.
References nnp::Dataset::Dataset(), epoch, forceWeight, freeMemory, hasStructures, hasUpdaters, jacobianMode, JM_SUM, numEpochs, numUpdaters, numWeights, parallelMode, PM_TRAIN_RK0, repeatedEnergyUpdates, stage, sw, trainingLogFileName, updaterType, updateStrategy, US_COMBINED, useForces, UT_GD, writeNeuronStatisticsAlways, writeNeuronStatisticsEvery, writeTrainingLog, writeWeightsAlways, and writeWeightsEvery.

◆ ~Training()
| Training::~Training | ( | ) |
Destructor, updater vector needs to be cleaned.
Definition at line 64 of file Training.cpp.
References trainingLog, updaters, updaterType, UT_GD, and UT_KF.
Member Function Documentation
◆ selectSets()
| void Training::selectSets | ( | ) |
Randomly select training and test set structures.
Also fills training candidates lists.
Definition at line 82 of file Training.cpp.
References nnp::Structure::atoms, nnp::Dataset::comm, nnp::Mode::elementMap, hasStructures, nnp::Mode::HDNNP_4G, nnp::Structure::index, nnp::Mode::log, MPI_SIZE_T, nnp::Dataset::myRank, nnp::Mode::nnpType, nnp::Structure::numAtoms, nnp::Structure::numAtomsPerElement, nnp::Mode::numElements, p, pk, nnp::Dataset::rng, nnp::Structure::sampleType, nnp::Structure::ST_TEST, nnp::Structure::ST_TRAINING, nnp::Structure::ST_UNKNOWN, nnp::strpr(), and nnp::Dataset::structures.
Referenced by main().

◆ writeSetsToFiles()
| void Training::writeSetsToFiles | ( | ) |
Write training and test set to separate files (train.data and test.data, same format as input.data).
Definition at line 230 of file Training.cpp.
References nnp::Mode::addEnergyOffset(), nnp::Dataset::combineFiles(), nnp::Dataset::comm, nnp::Mode::log, nnp::Dataset::myRank, nnp::Mode::removeEnergyOffset(), nnp::Structure::ST_TEST, nnp::Structure::ST_TRAINING, nnp::strpr(), and nnp::Dataset::structures.
Referenced by main().
◆ initializeWeights()
| void Training::initializeWeights | ( | ) |
Initialize weights for all elements.
Definition at line 292 of file Training.cpp.
References nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Mode::NNSetup::keywordSuffix2, nnp::Mode::log, nnp::Mode::NNSetup::name, nnId, nnp::Mode::nnpType, nnp::Mode::nns, randomizeNeuralNetworkWeights(), nnp::Mode::readNeuralNetworkWeights(), stage, and nnp::Mode::NNSetup::weightFileFormat.
Referenced by main().


◆ initializeWeightsMemory()
| void Training::initializeWeightsMemory | ( | UpdateStrategy | updateStrategy = US_COMBINED | ) |
Initialize weights vector according to update strategy.
- Parameters
-
[in] updateStrategy Determines the shape of the weights array.
Definition at line 330 of file Training.cpp.
References nnp::Mode::elements, nnp::Mode::HDNNP_4G, nnp::Mode::log, nnId, nnp::Mode::nnpType, nnp::Mode::numElements, numUpdaters, numWeights, numWeightsPerUpdater, stage, nnp::strpr(), updateStrategy, US_COMBINED, US_ELEMENT, weights, and weightsOffset.
Referenced by setupNumericDerivCheck(), and setupTraining().
◆ setStage()
| void Training::setStage | ( | std::size_t | stage | ) |
Set training stage (if multiple stages are needed for NNP type).
- Parameters
-
[in] stage Training stage to set.
Definition at line 379 of file Training.cpp.
References nnp::Mode::HDNNP_2G, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnId, nnp::Mode::nnpType, p, pk, stage, and nnp::strpr().
Referenced by main().


◆ dataSetNormalization()
| void Training::dataSetNormalization | ( | ) |
Apply normalization based on initial weights prediction.
Definition at line 429 of file Training.cpp.
References nnp::appendLinesToFile(), nnp::Mode::calculateAtomicNeuralNetworks(), nnp::Mode::calculateEnergy(), nnp::Mode::calculateForces(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), nnp::Dataset::combineFiles(), nnp::Mode::convEnergy, nnp::Mode::convLength, nnp::createFileHeader(), nnp::Mode::elements, nnp::Mode::getEnergyWithOffset(), nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Mode::log, nnp::Mode::maxCutoffRadius, nnp::Mode::meanEnergy, MPI_SIZE_T, nnp::Dataset::myRank, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Dataset::numStructures, nnp::Mode::setupNormalization(), nnp::Mode::setupSymmetryFunctionCache(), nnp::Mode::setupSymmetryFunctionGroups(), nnp::Mode::setupSymmetryFunctionMemory(), nnp::Mode::setupSymmetryFunctions(), nnp::Mode::setupSymmetryFunctionScaling(), nnp::Mode::setupSymmetryFunctionStatistics(), nnp::split(), stage, nnp::strpr(), nnp::Dataset::structures, nnp::Mode::writeSettingsFile(), and writeWeights().
Referenced by main().

◆ setupTraining()
| void Training::setupTraining | ( | ) |
General training settings and setup of weight update routine.
Definition at line 737 of file Training.cpp.
References allocateArrays(), nnp::appendLinesToFile(), nnp::createFileHeader(), nnp::GradientDescent::DT_ADAM, nnp::GradientDescent::DT_FIXED, nnp::Mode::elements, forceWeight, freeMemory, getWeights(), hasUpdaters, nnp::Mode::HDNNP_2G, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, initializeWeightsMemory(), jacobianMode, JM_FULL, JM_SUM, JM_TASK, nnp::KalmanFilter::KT_FADINGMEMORY, nnp::KalmanFilter::KT_STANDARD, nnp::Mode::log, nnp::Dataset::myRank, nnId, nnp::Mode::nnpType, nnp::Mode::nns, numEpochs, numUpdaters, nnp::Training::Property::numUpdates, numWeightsPerUpdater, p, parallelMode, nnp::Training::Property::patternsPerUpdateGlobal, pk, PM_TRAIN_ALL, PM_TRAIN_RK0, repeatedEnergyUpdates, nnp::Dataset::rng, nnp::Dataset::rngGlobal, rngGlobalNew, rngNew, nnp::GradientDescent::setParametersAdam(), nnp::KalmanFilter::setParametersFadingMemory(), nnp::GradientDescent::setParametersFixed(), nnp::KalmanFilter::setParametersStandard(), setupFileOutput(), setupSelectionMode(), setupUpdatePlan(), stage, nnp::strpr(), sw, trainingLog, trainingLogFileName, updaters, updaterType, updateStrategy, US_COMBINED, US_ELEMENT, useForces, UT_GD, UT_KF, UT_LM, weights, and writeTrainingLog.
Referenced by main().

◆ setupNumericDerivCheck()
| vector< string > Training::setupNumericDerivCheck | ( | ) |
Set up numeric weight derivatives check.
Definition at line 1209 of file Training.cpp.
References getWeights(), nnp::Mode::HDNNP_2G, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, initializeWeightsMemory(), nnp::Mode::log, nnId, nnp::Mode::nnpType, p, pk, nnp::Mode::readNeuralNetworkWeights(), and stage.
Referenced by main().

◆ calculateNeighborLists()
| void Training::calculateNeighborLists | ( | ) |
Calculate neighbor lists for all structures.
Definition at line 1247 of file Training.cpp.
References nnp::Mode::convLength, nnp::Mode::cutoffs, nnp::Mode::ewaldSetup, nnp::Mode::HDNNP_4G, nnp::Mode::log, nnp::Mode::maxCutoffRadius, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Mode::screeningFunction, nnp::strpr(), nnp::Dataset::structures, and sw.
Referenced by main().

◆ calculateError()
| void Training::calculateError | ( | std::map< std::string, std::pair< std::string, std::string > > const | fileNames | ) |
Calculate error metrics for all structures.
- Parameters
-
[in] fileNames Map of properties to file names for training/test comparison files.
If fileNames map is empty, no files will be written.
Definition at line 1310 of file Training.cpp.
References nnp::appendLinesToFile(), nnp::Mode::calculateAtomicNeuralNetworks(), nnp::Mode::calculateEnergy(), nnp::Mode::calculateForces(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), nnp::Mode::chargeEquilibration(), nnp::Dataset::collectError(), nnp::Dataset::combineFiles(), nnp::Dataset::comm, nnp::createFileHeader(), freeMemory, nnp::Mode::HDNNP_4G, nnp::Mode::maxCutoffRadius, nnp::Dataset::myRank, nnId, nnp::Mode::nnpType, p, pk, nnp::Structure::ST_TEST, nnp::Structure::ST_TRAINING, stage, nnp::strpr(), nnp::Dataset::structures, and useForces.
Referenced by calculateErrorEpoch().
◆ calculateErrorEpoch()
| void Training::calculateErrorEpoch | ( | ) |
Calculate error metrics per epoch for all structures with file names used in training loop.
Also write training curve to file.
Definition at line 1515 of file Training.cpp.
References calculateError(), d, epoch, p, and nnp::strpr().
Referenced by loop().

◆ calculateChargeErrorVec()
| void Training::calculateChargeErrorVec | ( | Structure const & | s, |
| Eigen::VectorXd & | cVec, | ||
| double & | cNorm ) |
Calculate vector of charge errors in one structure.
- Parameters
-
[in] s Structure of interest. [in] cVec Vector to store result of charge errors. [in] cNorm Euclidean norm of the error vector.
Definition at line 1545 of file Training.cpp.
References nnp::Structure::atoms, and nnp::Structure::numAtoms.
Referenced by update().
◆ printHeader()
| void Training::printHeader | ( | ) |
Print training loop header on screen.
Definition at line 1558 of file Training.cpp.
References nnp::Mode::log, p, pk, and nnp::strpr().
Referenced by loop().


◆ printEpoch()
| void Training::printEpoch | ( | ) |
Print preferred error metric and timing information on screen.
Definition at line 1616 of file Training.cpp.
References countUpdates, epoch, nnp::Mode::log, nnp::Mode::normalize, p, nnp::Mode::physical(), pk, nnp::strpr(), and sw.
Referenced by loop().

◆ writeWeights()
| void Training::writeWeights | ( | std::string const & | nnName, |
| std::string const & | fileNameFormat ) const |
Write weights to files (one file for each element).
- Parameters
-
[in] nnName Identifier for neural network. [in] fileNameFormat String with file name format.
Definition at line 1662 of file Training.cpp.
References nnp::Mode::elements, nnp::Mode::numElements, and nnp::strpr().
Referenced by dataSetNormalization(), main(), and writeWeightsEpoch().

◆ writeWeightsEpoch()
| void Training::writeWeightsEpoch | ( | ) | const |
Write weights to files during training loop.
Definition at line 1679 of file Training.cpp.
References epoch, nnp::Mode::HDNNP_2G, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnId, nnp::Mode::nnpType, stage, nnp::strpr(), writeWeights(), writeWeightsAlways, and writeWeightsEvery.
Referenced by loop().

◆ writeHardness()
| void Training::writeHardness | ( | std::string const & | fileNameFormat | ) | const |
Write hardness to files (one file for each element).
- Parameters
-
[in] fileNameFormat String with file name format.
Definition at line 1702 of file Training.cpp.
References nnp::Mode::elements, nnp::Mode::normalize, nnp::Mode::numElements, nnp::Mode::physical(), and nnp::strpr().
Referenced by writeHardnessEpoch().
◆ writeHardnessEpoch()
| void Training::writeHardnessEpoch | ( | ) | const |
Write hardness to files during training loop.
Definition at line 1720 of file Training.cpp.
References epoch, nnp::Mode::HDNNP_4G, nnp::Mode::nnpType, stage, nnp::strpr(), writeHardness(), writeWeightsAlways, and writeWeightsEvery.
Referenced by loop().

◆ writeLearningCurve()
| void Training::writeLearningCurve | ( | bool | append, |
| std::string const | fileName = "learning-curve.out" ) const |
Write current RMSEs and epoch information to file.
- Parameters
-
[in] append If true, append to file, otherwise create new file. [in] fileName File name for learning curve file.
Definition at line 1734 of file Training.cpp.
References nnp::appendLinesToFile(), nnp::createFileHeader(), epoch, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Mode::nnpType, nnp::Mode::normalize, p, nnp::Mode::physical(), pk, stage, and nnp::strpr().
Referenced by loop().
◆ writeNeuronStatistics()
| void Training::writeNeuronStatistics | ( | std::string const & | nnName, |
| std::string const & | fileName ) const |
Write neuron statistics collected since last invocation.
- Parameters
-
[in] nnName Identifier of neural network to process. [in] fileName File name for statistics file.
Definition at line 1844 of file Training.cpp.
References nnp::appendLinesToFile(), nnp::Dataset::comm, nnp::createFileHeader(), nnp::Mode::elements, nnp::Dataset::myRank, nnp::Mode::numElements, and nnp::strpr().
Referenced by writeNeuronStatisticsEpoch().
◆ writeNeuronStatisticsEpoch()
| void Training::writeNeuronStatisticsEpoch | ( | ) | const |
Write neuron statistics during training loop.
Definition at line 1943 of file Training.cpp.
References epoch, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnId, nnp::Mode::nnpType, stage, nnp::strpr(), writeNeuronStatistics(), writeNeuronStatisticsAlways, and writeNeuronStatisticsEvery.
Referenced by loop().
◆ resetNeuronStatistics()
| void Training::resetNeuronStatistics | ( | ) |
Reset neuron statistics for all elements.
Definition at line 1962 of file Training.cpp.
References nnp::Mode::elements.
Referenced by loop().
◆ writeUpdaterStatus()
| void Training::writeUpdaterStatus | ( | bool | append, |
| std::string const | fileNameFormat = "updater.%03zu.out" ) const |
Write updater information to file.
- Parameters
-
[in] append If true, append to file, otherwise create new file. [in] fileNameFormat String with file name format.
Definition at line 1972 of file Training.cpp.
References nnp::appendLinesToFile(), nnp::Mode::elementMap, epoch, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Mode::nnpType, numUpdaters, stage, nnp::strpr(), updaters, updateStrategy, US_COMBINED, and US_ELEMENT.
Referenced by loop().
◆ sortUpdateCandidates()
| void Training::sortUpdateCandidates | ( | std::string const & | property | ) |
Sort update candidates with descending RMSE.
- Parameters
-
[in] property Training property.
Definition at line 2008 of file Training.cpp.
References nnp::Structure::atoms, nnp::Structure::energy, nnp::Structure::energyRef, nnp::Atom::f, nnp::Atom::fRef, nnp::Structure::numAtoms, p, and nnp::Dataset::structures.
Referenced by loop().
◆ shuffleUpdateCandidates()
| void Training::shuffleUpdateCandidates | ( | std::string const & | property | ) |
Shuffle update candidates.
- Parameters
-
[in] property Training property.
Definition at line 2062 of file Training.cpp.
Referenced by loop().
◆ checkSelectionMode()
| void Training::checkSelectionMode | ( | ) |
Check if selection mode should be changed.
Definition at line 2114 of file Training.cpp.
References epoch, nnp::Mode::log, p, pk, SM_RANDOM, SM_SORT, SM_THRESHOLD, and nnp::strpr().
Referenced by loop().


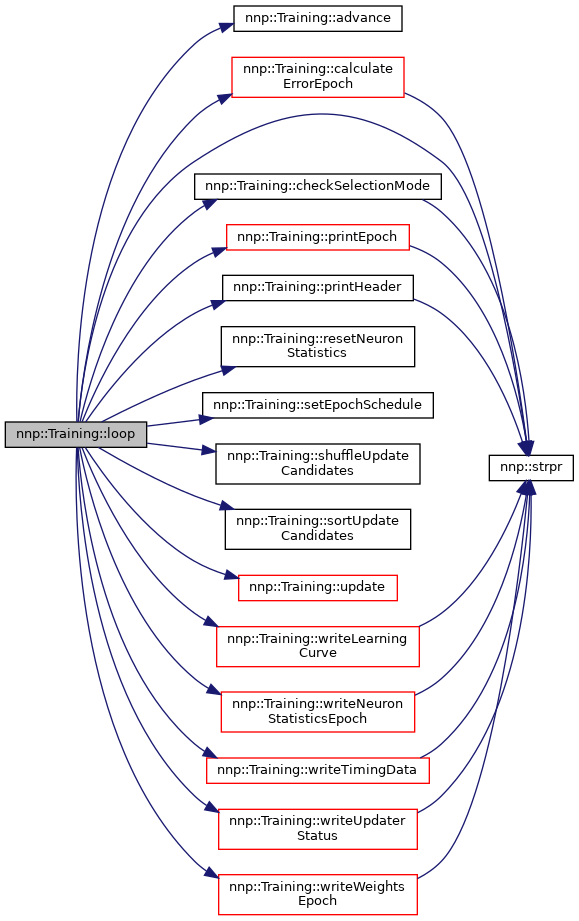
◆ loop()
| void Training::loop | ( | ) |
Execute main training loop.
Definition at line 2147 of file Training.cpp.
References advance(), calculateErrorEpoch(), checkSelectionMode(), epoch, epochSchedule, nnp::Mode::log, nnp::Dataset::myRank, p, pk, printEpoch(), printHeader(), resetNeuronStatistics(), setEpochSchedule(), shuffleUpdateCandidates(), SM_SORT, sortUpdateCandidates(), nnp::strpr(), sw, update(), writeHardnessEpoch(), writeLearningCurve(), writeNeuronStatisticsEpoch(), writeTimingData(), writeUpdaterStatus(), and writeWeightsEpoch().
Referenced by main().

◆ setEpochSchedule()
| void Training::setEpochSchedule | ( | ) |
Select energies/forces schedule for one epoch.
Definition at line 2086 of file Training.cpp.
References epochSchedule, p, pk, and rngGlobalNew.
Referenced by loop().

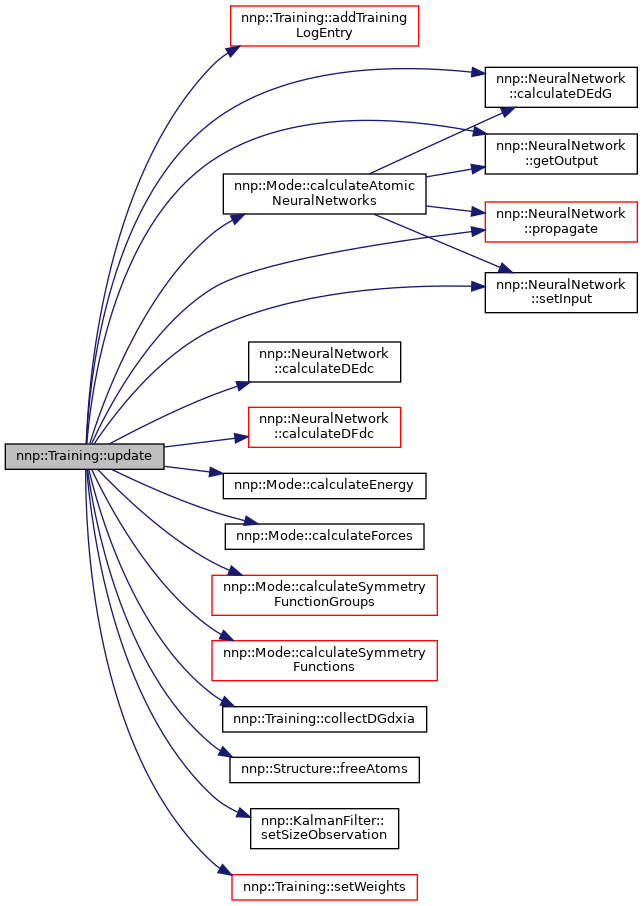
◆ update()
| void Training::update | ( | std::string const & | property | ) |
Perform one update.
- Parameters
-
[in] property Training property to use for update.
Definition at line 2256 of file Training.cpp.
References nnp::Training::SubCandidate::a, addTrainingLogEntry(), nnp::Structure::atoms, nnp::Training::SubCandidate::c, nnp::Mode::calculateAtomicNeuralNetworks(), calculateChargeErrorVec(), nnp::NeuralNetwork::calculateDEdc(), nnp::NeuralNetwork::calculateDEdG(), nnp::NeuralNetwork::calculateDFdc(), nnp::Structure::calculateDQdChi(), nnp::Structure::calculateDQdJ(), nnp::Structure::calculateDQdr(), nnp::Mode::calculateEnergy(), nnp::Mode::calculateForces(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), nnp::Atom::charge, nnp::Mode::chargeEquilibration(), nnp::Atom::chargeRef, nnp::Atom::chi, nnp::Structure::clearElectrostatics(), collectDGdxia(), nnp::Dataset::comm, nnp::Training::Property::countGroupedSubCand, countUpdates, nnp::Training::Property::countUpdates, nnp::Atom::dChidG, dGdxia, nnp::Atom::element, nnp::Mode::elements, nnp::Structure::energy, nnp::Structure::energyRef, nnp::Training::Property::epochFraction, nnp::Training::Property::error, nnp::Training::Property::errorsPerTask, nnp::Training::Property::errorTrain, nnp::Atom::f, forceWeight, nnp::Structure::freeAtoms(), freeMemory, nnp::Atom::fRef, nnp::Atom::G, nnp::NeuralNetwork::getOutput(), nnp::Structure::hasAMatrix, nnp::Structure::hasCharges, nnp::Mode::HDNNP_2G, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Training::Property::jacobian, jacobianMode, JM_FULL, JM_SUM, JM_TASK, nnp::Mode::maxCutoffRadius, MPI_SIZE_T, nnp::Dataset::myRank, nnId, nnp::Mode::nnpType, nnp::Mode::normalize, nnp::Mode::normalized(), nnp::Structure::numAtoms, nnp::Structure::numAtomsPerElement, nnp::Mode::numElements, nnp::Training::Property::numGroupedSubCand, nnp::Atom::numNeighbors, nnp::Atom::numNeighborsPerElement, nnp::Dataset::numProcs, numUpdaters, numWeightsPerUpdater, nnp::Training::Property::offsetJacobian, nnp::Training::Property::offsetPerTask, p, parallelMode, nnp::Training::Property::patternsPerUpdate, PM_TRAIN_ALL, PM_TRAIN_RK0, nnp::Training::UpdateCandidate::posSubCandidates, nnp::Training::Property::posUpdateCandidates, nnp::NeuralNetwork::propagate(), nnp::Training::Property::rmseThreshold, nnp::Training::Property::rmseThresholdTrials, nnp::Training::UpdateCandidate::s, nnp::Training::Property::selectionMode, nnp::NeuralNetwork::setInput(), nnp::KalmanFilter::setSizeObservation(), setWeights(), SM_RANDOM, SM_SORT, SM_THRESHOLD, nnp::Dataset::structures, nnp::Training::UpdateCandidate::subCandidates, sw, nnp::Training::Property::taskBatchSize, nnp::Training::Property::updateCandidates, updaters, updaterType, updateStrategy, US_COMBINED, US_ELEMENT, UT_KF, weights, weightsOffset, nnp::Training::Property::weightsPerTask, and writeTrainingLog.
Referenced by loop().
◆ getSingleWeight()
| double Training::getSingleWeight | ( | std::size_t | element, |
| std::size_t | index ) |
Get a single weight value.
- Parameters
-
[in] element Element index of weight. [in] index Weight index.
- Returns
- Weight value.
Note: This function is implemented for testing purposes and works correctly only with update strategy US_ELEMENT.
Definition at line 3158 of file Training.cpp.
References getWeights(), and weights.
◆ setSingleWeight()
| void Training::setSingleWeight | ( | std::size_t | element, |
| std::size_t | index, | ||
| double | value ) |
Set a single weight value.
- Parameters
-
[in] element Element index of weight. [in] index Weight index. [in] value Weight value.
Note: This function is implemented for testing purposes and works correctly only with update strategy US_ELEMENT.
Definition at line 3165 of file Training.cpp.
References setWeights(), and weights.
◆ calculateWeightDerivatives() [1/2]
| vector< vector< double > > Training::calculateWeightDerivatives | ( | Structure * | structure | ) |
Calculate derivatives of energy with respect to weights.
- Parameters
-
[in,out] structure Structure to process.
- Returns
- Vector with derivatives of energy with respect to weights (per element).
- Note
- This function is implemented for testing purposes.
Definition at line 3174 of file Training.cpp.
References nnp::Structure::atoms, nnp::NeuralNetwork::calculateDEdc(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), nnp::Mode::elements, nnp::NeuralNetwork::getOutput(), nnp::Mode::numElements, nnp::NeuralNetwork::propagate(), and nnp::NeuralNetwork::setInput().
◆ calculateWeightDerivatives() [2/2]
| vector< vector< double > > Training::calculateWeightDerivatives | ( | Structure * | structure, |
| std::size_t | atom, | ||
| std::size_t | component ) |
Calculate derivatives of force with respect to weights.
- Parameters
-
[in,out] structure Structure to process. [in] atom Atom index. [in] component x, y or z-component of force (0, 1, 2).
- Returns
- Vector with derivatives of force with respect to weights (per element).
- Note
- This function is implemented for testing purposes.
Definition at line 3214 of file Training.cpp.
References nnp::Structure::atoms, nnp::NeuralNetwork::calculateDFdc(), nnp::Mode::calculateSymmetryFunctionGroups(), nnp::Mode::calculateSymmetryFunctions(), collectDGdxia(), dGdxia, nnp::Mode::elements, nnp::NeuralNetwork::getOutput(), nnp::Mode::maxCutoffRadius, nnp::Mode::numElements, nnp::NeuralNetwork::propagate(), and nnp::NeuralNetwork::setInput().
◆ setTrainingLogFileName()
| void Training::setTrainingLogFileName | ( | std::string | fileName | ) |
Set training log file name.
- Parameters
-
[in] fileName File name for training log.
Definition at line 3265 of file Training.cpp.
References trainingLogFileName.
◆ getNumConnections()
| size_t Training::getNumConnections | ( | std::string | id = "short" | ) | const |
Get total number of NN connections.
- Parameters
-
[in] id NN ID to use (e.g. "short").
- Returns
- Sum of all NN weights + biases for all elements
Definition at line 3272 of file Training.cpp.
References nnp::Mode::elements.
Referenced by dPdc().
◆ getNumConnectionsPerElement()
| vector< size_t > Training::getNumConnectionsPerElement | ( | std::string | id = "short" | ) | const |
Get number of NN connections for each element.
- Parameters
-
[in] id NN ID to use (e.g. "short").
- Returns
- Vector containing number of connections for each element.
Definition at line 3283 of file Training.cpp.
References nnp::Mode::elements.
Referenced by dPdc(), and dPdcN().
◆ getConnectionOffsets()
| vector< size_t > Training::getConnectionOffsets | ( | std::string | id = "short" | ) | const |
Get offsets of NN connections for each element.
- Parameters
-
[in] id NN ID to use (e.g. "short").
- Returns
- Vector containing offsets for each element.
Definition at line 3294 of file Training.cpp.
References nnp::Mode::elements.
Referenced by dPdc(), and dPdcN().

◆ dPdc()
| void Training::dPdc | ( | std::string | property, |
| Structure & | structure, | ||
| std::vector< std::vector< double > > & | dEdc ) |
Compute derivatives of property with respect to weights.
- Parameters
-
[in] property Training property for which derivatives should be computed. [in] structure The structure under investigation. [in,out] dEdc Weight derivative array (first index = property, second index = weight).
Definition at line 3307 of file Training.cpp.
References nnp::Structure::atoms, nnp::NeuralNetwork::calculateDEdc(), nnp::NeuralNetwork::calculateDEdG(), nnp::NeuralNetwork::calculateDFdc(), collectDGdxia(), dGdxia, dPdc(), nnp::Mode::elements, getConnectionOffsets(), getNumConnections(), getNumConnectionsPerElement(), nnp::NeuralNetwork::getOutput(), nnId, nnp::Structure::numAtoms, nnp::NeuralNetwork::propagate(), and nnp::NeuralNetwork::setInput().
Referenced by dPdc(), dPdcN(), and main().
◆ dPdcN()
| void Training::dPdcN | ( | std::string | property, |
| Structure & | structure, | ||
| std::vector< std::vector< double > > & | dEdc, | ||
| double | delta = 1.0E-4 ) |
Compute numeric derivatives of property with respect to weights.
- Parameters
-
[in] property Training property for which derivatives should be computed. [in] structure The structure under investigation. [in,out] dEdc Weight derivative array (first index = property, second index = weight). [in] delta Delta for central difference.
Definition at line 3379 of file Training.cpp.
References nnp::Structure::atoms, nnp::Mode::calculateAtomicNeuralNetworks(), nnp::Mode::calculateEnergy(), nnp::Mode::calculateForces(), dPdc(), nnp::Structure::energy, getConnectionOffsets(), getNumConnectionsPerElement(), nnp::Structure::numAtoms, nnp::Mode::numElements, setWeights(), and weights.
Referenced by main().

◆ advance()
|
private |
Check if training loop should be continued.
- Returns
- True if further training should be performed, false otherwise.
Definition at line 3461 of file Training.cpp.
References epoch, and numEpochs.
Referenced by loop().
◆ getWeights()
|
private |
Get weights from neural network class.
Definition at line 3467 of file Training.cpp.
References nnp::Mode::elements, nnp::NeuralNetwork::getConnections(), nnp::NeuralNetwork::getNumConnections(), nnp::Mode::HDNNP_4G, nnId, nnp::Mode::nnpType, nnp::Mode::numElements, stage, updateStrategy, US_COMBINED, US_ELEMENT, and weights.
Referenced by getSingleWeight(), setupNumericDerivCheck(), and setupTraining().
◆ setWeights()
|
private |
Set weights in neural network class.
Definition at line 3498 of file Training.cpp.
References nnp::Mode::elements, nnp::NeuralNetwork::getNumConnections(), nnp::Mode::HDNNP_4G, nnId, nnp::Mode::nnpType, nnp::Mode::numElements, nnp::NeuralNetwork::setConnections(), stage, updateStrategy, US_COMBINED, US_ELEMENT, and weights.
Referenced by dPdcN(), setSingleWeight(), and update().
◆ addTrainingLogEntry() [1/3]
|
private |
Write energy update data to training log file.
- Parameters
-
[in] proc Processor which provided update candidate. [in] il Loop index of threshold loop. [in] f RMSE fraction of update candidate. [in] is Local structure index. [in] isg Global structure index.
Definition at line 3529 of file Training.cpp.
References countUpdates, epoch, nnp::strpr(), and trainingLog.
Referenced by update().

◆ addTrainingLogEntry() [2/3]
|
private |
Write force update data to training log file.
- Parameters
-
[in] proc Processor which provided update candidate. [in] il Loop index of threshold loop. [in] f RMSE fraction of update candidate. [in] is Local structure index. [in] isg Global structure index. [in] ia Atom index. [in] ic Component index.
Definition at line 3543 of file Training.cpp.
References countUpdates, epoch, nnp::strpr(), and trainingLog.

◆ addTrainingLogEntry() [3/3]
|
private |
Write charge update data to training log file.
- Parameters
-
[in] proc Processor which provided update candidate. [in] il Loop index of threshold loop. [in] f RMSE fraction of update candidate. [in] is Local structure index. [in] isg Global structure index. [in] ia Atom index.
Definition at line 3559 of file Training.cpp.
References countUpdates, epoch, nnp::strpr(), and trainingLog.

◆ collectDGdxia()
|
private |
Collect derivative of symmetry functions with respect to one atom's coordinate.
- Parameters
-
[in] atom The atom which owns the symmetry functions. [in] indexAtom The index \(i\) of the atom requested. [in] indexComponent The component \(\alpha\) of the atom requested.
This calculates an array of derivatives
\[ \left(\frac{\partial G_1}{\partial x_{i,\alpha}}, \ldots, \frac{\partial G_n}{\partial x_{i,\alpha}}\right), \]
where \(\{G_j\}_{j=1,\ldots,n}\) are the symmetry functions for this atom and \(x_{i,\alpha}\) is the \(\alpha\)-component of the position of atom \(i\). The result is stored in dGdxia.
Definition at line 3574 of file Training.cpp.
References nnp::Atom::dGdr, nnp::Atom::Neighbor::dGdr, dGdxia, nnp::Atom::element, nnp::Atom::Neighbor::element, nnp::Mode::elements, nnp::Mode::HDNNP_4G, nnp::Atom::index, nnp::Atom::neighbors, nnp::Mode::nnpType, nnp::Atom::numNeighbors, and nnp::Atom::numSymmetryFunctions.
Referenced by calculateWeightDerivatives(), dPdc(), and update().
◆ randomizeNeuralNetworkWeights()
|
private |
Randomly initialize specificy neural network weights.
- Parameters
-
[in] id Actual network type to initialize ("short" or "elec").
Definition at line 3616 of file Training.cpp.
References nnp::Mode::elements, nnp::NeuralNetwork::getNumConnections(), nnp::Mode::log, nnp::NeuralNetwork::modifyConnections(), nnp::NeuralNetwork::MS_GLOROTBENGIO, nnp::NeuralNetwork::MS_NGUYENWIDROW, nnp::NeuralNetwork::MS_ZEROBIAS, nnp::Mode::nns, nnp::Mode::numElements, nnp::Dataset::rngGlobal, nnp::NeuralNetwork::setConnections(), and nnp::strpr().
Referenced by initializeWeights().
◆ setupSelectionMode()
|
private |
Set selection mode for specific training property.
- Parameters
-
[in] property Training property (uses corresponding keyword).
Definition at line 3669 of file Training.cpp.
References nnp::Mode::log, p, pk, SM_RANDOM, SM_SORT, SM_THRESHOLD, nnp::split(), and nnp::strpr().
Referenced by setupTraining().

◆ setupFileOutput()
|
private |
Set file output intervals for properties and other quantities.
- Parameters
-
[in] type Training property or weightsorneuron-stats.
Definition at line 3777 of file Training.cpp.
References nnp::Mode::log, p, pk, nnp::reduce(), nnp::split(), nnp::strpr(), writeNeuronStatisticsAlways, writeNeuronStatisticsEvery, writeWeightsAlways, and writeWeightsEvery.
Referenced by setupTraining().
◆ setupUpdatePlan()
|
private |
Set up how often properties are updated.
- Parameters
-
[in] property Training property (uses corresponding keyword).
Definition at line 3839 of file Training.cpp.
References nnp::Dataset::comm, nnp::Training::Property::epochFraction, nnp::Training::Property::errorsPerTask, jacobianMode, JM_FULL, nnp::Mode::log, MPI_SIZE_T, nnp::Dataset::myRank, nnp::Training::Property::numErrorsGlobal, nnp::Dataset::numProcs, nnp::Training::Property::numTrainPatterns, numUpdaters, nnp::Training::Property::numUpdates, numWeightsPerUpdater, nnp::Training::Property::offsetJacobian, nnp::Training::Property::offsetPerTask, p, nnp::Training::Property::patternsPerUpdate, nnp::Training::Property::patternsPerUpdateGlobal, pk, nnp::strpr(), nnp::Training::Property::taskBatchSize, nnp::Training::Property::updateCandidates, and nnp::Training::Property::weightsPerTask.
Referenced by setupTraining().

◆ allocateArrays()
|
private |
Allocate error and Jacobian arrays for given property.
- Parameters
-
[in] property Training property.
Definition at line 3949 of file Training.cpp.
References nnp::Training::Property::error, nnp::Training::Property::errorsPerTask, nnp::Training::Property::jacobian, jacobianMode, JM_SUM, nnp::Mode::log, nnp::Dataset::myRank, nnp::Training::Property::numErrorsGlobal, numUpdaters, numWeightsPerUpdater, p, parallelMode, pk, PM_TRAIN_ALL, PM_TRAIN_RK0, and nnp::strpr().
Referenced by setupTraining().


◆ writeTimingData()
|
private |
Write timing data for all clocks.
- Parameters
-
[in] append If true, append to file, otherwise create new file. [in] fileName File name for timing data file.
Definition at line 3987 of file Training.cpp.
References nnp::appendLinesToFile(), nnp::createFileHeader(), epoch, nnp::Mode::HDNNP_4G, nnp::Mode::HDNNP_Q, nnp::Mode::nnpType, p, pk, stage, nnp::strpr(), and sw.
Referenced by loop().

Member Data Documentation
◆ updaterType
|
private |
Updater type used.
Definition at line 503 of file Training.h.
Referenced by setupTraining(), Training(), update(), and ~Training().
◆ parallelMode
|
private |
Parallelization mode used.
Definition at line 505 of file Training.h.
Referenced by allocateArrays(), setupTraining(), Training(), and update().
◆ jacobianMode
|
private |
Jacobian mode used.
Definition at line 507 of file Training.h.
Referenced by allocateArrays(), setupTraining(), setupUpdatePlan(), Training(), and update().
◆ updateStrategy
|
private |
Update strategy used.
Definition at line 509 of file Training.h.
Referenced by getWeights(), initializeWeightsMemory(), setupTraining(), setWeights(), Training(), update(), and writeUpdaterStatus().
◆ hasUpdaters
|
private |
If this rank performs weight updates.
Definition at line 511 of file Training.h.
Referenced by setupTraining(), and Training().
◆ hasStructures
|
private |
If this rank holds structure information.
Definition at line 513 of file Training.h.
Referenced by selectSets(), and Training().
◆ useForces
|
private |
Use forces for training.
Definition at line 515 of file Training.h.
Referenced by calculateError(), setupTraining(), and Training().
◆ repeatedEnergyUpdates
|
private |
After force update perform energy update for corresponding structure.
Definition at line 517 of file Training.h.
Referenced by setupTraining(), and Training().
◆ freeMemory
|
private |
Free symmetry function memory after calculation.
Definition at line 519 of file Training.h.
Referenced by calculateError(), setupTraining(), Training(), and update().
◆ writeTrainingLog
|
private |
Whether training log file is written.
Definition at line 521 of file Training.h.
Referenced by setupTraining(), Training(), and update().
◆ stage
|
private |
Training stage.
Definition at line 523 of file Training.h.
Referenced by calculateError(), dataSetNormalization(), getWeights(), initializeWeights(), initializeWeightsMemory(), setStage(), setupNumericDerivCheck(), setupTraining(), setWeights(), Training(), writeHardnessEpoch(), writeLearningCurve(), writeNeuronStatisticsEpoch(), writeTimingData(), writeUpdaterStatus(), and writeWeightsEpoch().
◆ numUpdaters
|
private |
Number of updaters (depends on update strategy).
Definition at line 525 of file Training.h.
Referenced by allocateArrays(), initializeWeightsMemory(), setupTraining(), setupUpdatePlan(), Training(), update(), and writeUpdaterStatus().
◆ numEpochs
|
private |
Number of epochs requested.
Definition at line 527 of file Training.h.
Referenced by advance(), setupTraining(), and Training().
◆ epoch
|
private |
Current epoch.
Definition at line 529 of file Training.h.
Referenced by addTrainingLogEntry(), addTrainingLogEntry(), addTrainingLogEntry(), advance(), calculateErrorEpoch(), checkSelectionMode(), loop(), printEpoch(), Training(), writeHardnessEpoch(), writeLearningCurve(), writeNeuronStatisticsEpoch(), writeTimingData(), writeUpdaterStatus(), and writeWeightsEpoch().
◆ writeWeightsEvery
|
private |
Write weights every this many epochs.
Definition at line 531 of file Training.h.
Referenced by setupFileOutput(), Training(), writeHardnessEpoch(), and writeWeightsEpoch().
◆ writeWeightsAlways
|
private |
Up to this epoch weights are written every epoch.
Definition at line 533 of file Training.h.
Referenced by setupFileOutput(), Training(), writeHardnessEpoch(), and writeWeightsEpoch().
◆ writeNeuronStatisticsEvery
|
private |
Write neuron statistics every this many epochs.
Definition at line 535 of file Training.h.
Referenced by setupFileOutput(), Training(), and writeNeuronStatisticsEpoch().
◆ writeNeuronStatisticsAlways
|
private |
Up to this epoch neuron statistics are written every epoch.
Definition at line 537 of file Training.h.
Referenced by setupFileOutput(), Training(), and writeNeuronStatisticsEpoch().
◆ countUpdates
|
private |
Update counter (for all training quantities together).
Definition at line 539 of file Training.h.
Referenced by addTrainingLogEntry(), addTrainingLogEntry(), addTrainingLogEntry(), printEpoch(), and update().
◆ numWeights
|
private |
Total number of weights.
If nnpType 4G also h = sqrt(J) (hardness) is counted.
Definition at line 542 of file Training.h.
Referenced by initializeWeightsMemory(), and Training().
◆ forceWeight
|
private |
Force update weight.
Definition at line 544 of file Training.h.
Referenced by setupTraining(), Training(), and update().
◆ trainingLogFileName
|
private |
File name for training log.
Definition at line 546 of file Training.h.
Referenced by setTrainingLogFileName(), setupTraining(), and Training().
◆ nnId
|
private |
ID of neural network the training is working on.
Definition at line 548 of file Training.h.
Referenced by calculateError(), dPdc(), getWeights(), initializeWeights(), initializeWeightsMemory(), setStage(), setupNumericDerivCheck(), setupTraining(), setWeights(), update(), writeNeuronStatisticsEpoch(), and writeWeightsEpoch().
◆ trainingLog
|
private |
Training log file.
Definition at line 550 of file Training.h.
Referenced by addTrainingLogEntry(), addTrainingLogEntry(), addTrainingLogEntry(), setupTraining(), and ~Training().
◆ epochSchedule
|
private |
Update schedule epoch (stage 1: 0 = charge update; stage 2: 0 = energy update, 1 = force update (optional)).
Definition at line 553 of file Training.h.
Referenced by loop(), and setEpochSchedule().
◆ numWeightsPerUpdater
|
private |
Number of weights per updater.
If nnpType 4G also h = sqrt(J) is counted during stage 1 training.
Definition at line 556 of file Training.h.
Referenced by allocateArrays(), initializeWeightsMemory(), setupTraining(), setupUpdatePlan(), and update().
◆ weightsOffset
|
private |
Offset of each element's weights in combined array.
If nnpType 4G also h = sqrt(J) is counted.
Definition at line 559 of file Training.h.
Referenced by initializeWeightsMemory(), and update().
◆ pk
|
private |
Vector of actually used training properties.
Definition at line 561 of file Training.h.
Referenced by allocateArrays(), calculateError(), checkSelectionMode(), loop(), printEpoch(), printHeader(), selectSets(), setEpochSchedule(), setStage(), setupFileOutput(), setupNumericDerivCheck(), setupSelectionMode(), setupTraining(), setupUpdatePlan(), writeLearningCurve(), and writeTimingData().
◆ dGdxia
|
private |
Derivative of symmetry functions with respect to one specific atom coordinate.
Definition at line 565 of file Training.h.
Referenced by calculateWeightDerivatives(), collectDGdxia(), dPdc(), and update().
◆ weights
|
private |
Neural network weights and biases for each element.
If nnpType 4G also h = sqrt(J) included.
Definition at line 570 of file Training.h.
Referenced by dPdcN(), getSingleWeight(), getWeights(), initializeWeightsMemory(), setSingleWeight(), setupTraining(), setWeights(), and update().
◆ updaters
|
private |
Weight updater (combined or for each element).
Definition at line 572 of file Training.h.
Referenced by setupTraining(), update(), writeUpdaterStatus(), and ~Training().
◆ sw
|
private |
Stopwatches for timing overview.
Definition at line 575 of file Training.h.
Referenced by calculateNeighborLists(), loop(), printEpoch(), setupTraining(), Training(), update(), and writeTimingData().
◆ rngNew
|
private |
Per-task random number generator.
Definition at line 577 of file Training.h.
Referenced by setupTraining(), and shuffleUpdateCandidates().
◆ rngGlobalNew
|
private |
Global random number generator.
Definition at line 579 of file Training.h.
Referenced by setEpochSchedule(), and setupTraining().
◆ p
|
private |
Actual training properties.
Definition at line 581 of file Training.h.
Referenced by allocateArrays(), calculateError(), calculateErrorEpoch(), checkSelectionMode(), loop(), printEpoch(), printHeader(), selectSets(), setEpochSchedule(), setStage(), setupFileOutput(), setupNumericDerivCheck(), setupSelectionMode(), setupTraining(), setupUpdatePlan(), shuffleUpdateCandidates(), sortUpdateCandidates(), update(), writeLearningCurve(), and writeTimingData().
The documentation for this class was generated from the following files:
- /home/runner/work/n2p2/n2p2/src/libnnptrain/Training.h
- /home/runner/work/n2p2/n2p2/src/libnnptrain/Training.cpp